|
Below are summaries of molecular phylogenetics/evolution/biogeography research projects that are currently in progress in the laboratory of Chris Simon (University of Connecticut And Victoria University of Wellington). Some of these are collaborations with researchers in other states and countries.
At the end of each description, you'll find a link for more information and a discussion of of potential new projects including both small and large studies.
Paternal inheritance of mitochondria
Chris Simon1, John Cooley1,2, Kathryn Fontaine1, Kashiwa Hereford1
1The University of Connecticut
2Yale University

 Animal mitochondrial DNA (mtDNA) is generally assumed to be maternally inherited. Is this assumption correct? In the last 20 years, several instances have been discovered in which animal mtDNA is transmitted through patrilines, a phenomenon termed "paternal leakage". It challenges some of the assumptions involved in using mtDNA as a molecular or forensic marker and it can complicate phylogenetic reconstruction and molecular dating. We have demonstrated paternal leakage in experimental hybrid crosses of periodical cicadas, and we have taken advantage of the phenomenon of paternal leakage to demonstrate that male periodical cicadas can gain from mating an already-mated female. These results provide important clues about why male periodical cicadas are so persistent in approaching and courting even unreceptive females. Read more about this project here.
Animal mitochondrial DNA (mtDNA) is generally assumed to be maternally inherited. Is this assumption correct? In the last 20 years, several instances have been discovered in which animal mtDNA is transmitted through patrilines, a phenomenon termed "paternal leakage". It challenges some of the assumptions involved in using mtDNA as a molecular or forensic marker and it can complicate phylogenetic reconstruction and molecular dating. We have demonstrated paternal leakage in experimental hybrid crosses of periodical cicadas, and we have taken advantage of the phenomenon of paternal leakage to demonstrate that male periodical cicadas can gain from mating an already-mated female. These results provide important clues about why male periodical cicadas are so persistent in approaching and courting even unreceptive females. Read more about this project here.
|
Pleistocene Forest Refugia in New Zealand
Thomas Buckley3, Maureen Mara4, Glenn Thackray5, Chris Simon1,2, David Marshall1, Kathy Hill1, Colleen Chambers1, and Richard Leschen3
1The University of Connecticut
2Victoria University, Wellington
3Landcare, Auckland
4The University of Canterbury
5Idaho State University
|
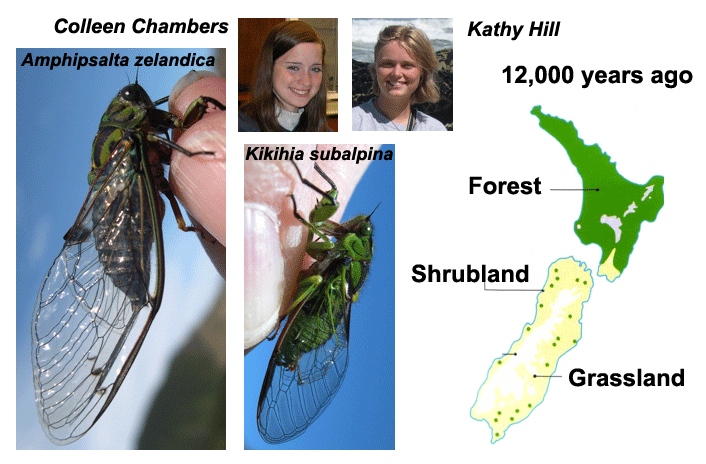
This Marsden Grant is a collaboration, funded in 2005. The Simon lab is sequencing two forest species of NZ cicadas and analyzing these data in conjunction with that of the other species described in the proposal. Summary: The Ice Ages caused massive changes in the distribution and composition of ecological communities. During the Last Glacial Maximum, or LGM (34,000 years to 17,650 years ago), much of the forest cover of New Zealand was replaced with a tundra-like environment. However, it is not clear if the forest survived only in large patches, known as refugia, in the North Island and the north of the South Island, or whether small refugia were scattered throughout the South Island. We will use genetic, climatic, palaeontological, and geological data to determine whether forest insect populations survived in these scattered refugia during the LGM or whether the modern populations are derived from the large forested areas further north. Specifically, we will apply population genetic analyses to ten widespread [forest-dwelling] insect species to detect the genetic signature of ancient refugia. We will also perform climate-modelling analyses on distributional data from these same insects and couple these data with information on glacial advances, and reconstructions of fossil insect ecosystems. Integrating these data will allow us to build a comprehensive description of the environment in the South Island in terms of the biogeography, ecology, climate and the response of taxa to LGM climatic fluctuations. The needed sequences for K. subalpina were collected by undergrad student Kathryn Gannon (now Fontaine) and technician Kathy Hill. Undergraduate student Colleen Chambers is collecting data for Amphipsalta zelandica and it relatives in NZ.
This project is nearing completion but results of other phylogeography projects on NZ cicadas listed above would interface nicely with this project.
|
Phylogenetics and Phylogeography of New Zealand Cicadas
Dave Marshall11, Kathy Hill1, John Cooley1, 4, Adam Leston1, Michael Cordiero1, Thomas Buckley3, and Chris Simon1,2
1The University of Connecticut
2Victoria University, Wellington
3Landcare, Auckland
4Yale University

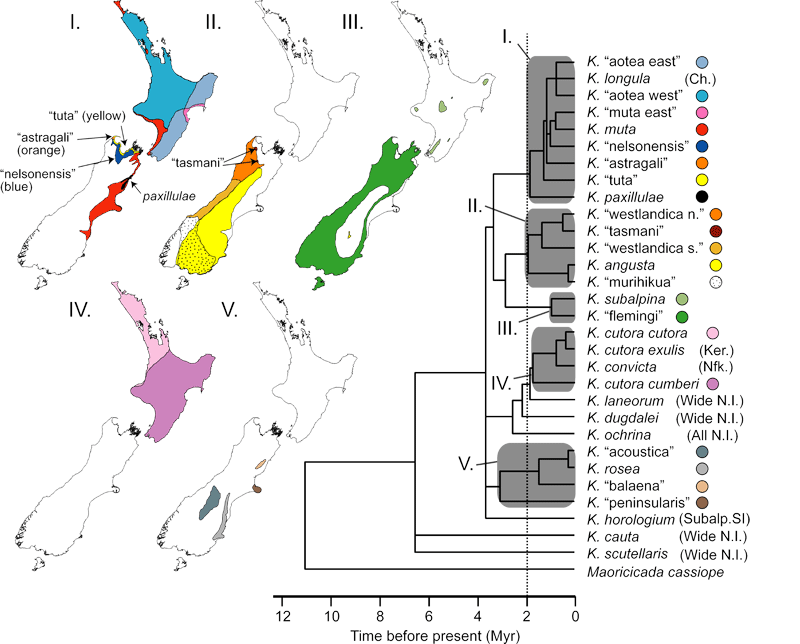 This project, funded by the U.S. National Science Foundation, addresses questions of how past changes in climate and land forms affect speciation. New combinations of phylogenetic and phylogeographic techniques using multiple gene loci are becoming key tools in the description of genetic diversity within species. These approaches can test detailed hypotheses of population and species origin, range changes, and extinction. We are conducting a fine-scale phylogeographic study of three widely co-distributed species complexes of New Zealand cicadas in three genera based on multiple independent nuclear introns and mitochondrial DNA: the Kikiha muta complex, the Maoricicada campbelli complex, and the Rhodopsalta species complex. Analyses of molecular data, in combination with song and morphological differences, will improve our understanding of how genetic differences among populations that are geographically isolated to varying degrees may eventually lead to speciation. It will also allow us to test a number of interesting biogeographic hypothesis based on the well-studied paleo-climatological and geological history of New Zealand. In the process we will gain a more complete picture of NZ cicada species diversity (broadly surveyed in the 1960s-70s). The islands of NZ are intriguing because they include a biota composed of both continental and island elements and a range of habitats as diverse as alpine tundra, sand dunes and subtropical to cold temperate rainforest. Our previous work suggests that cicadas arrived in NZ as two colonizing species--one with Australian affinities and one with New Caledonian affinities--about 10 m years ago and subsequently evolved into approximately 40-60 species occupying virtually all habitat types. Our approach of constructing phylogeographic trees for multiple independent nuclear and mitochondrial DNA data sets is necessary because species histories are complicated by such factors as hybridization and random inheritance of polymorphic ancestral genotypes that can only be recognized when independent genes are compared. We have created detailed phylogeographic trees for three NZ cicada species complexes. Each of these species groups has been sampled from many parts of the species range; multiple cryptic species have been discovered. Courtship songs have been recorded for a majority of the individuals sampled. Speciation events have been time calibrated using a variety of relaxed-molecular-clock methods and we have studied diversification rates through time. Publications include:
This project, funded by the U.S. National Science Foundation, addresses questions of how past changes in climate and land forms affect speciation. New combinations of phylogenetic and phylogeographic techniques using multiple gene loci are becoming key tools in the description of genetic diversity within species. These approaches can test detailed hypotheses of population and species origin, range changes, and extinction. We are conducting a fine-scale phylogeographic study of three widely co-distributed species complexes of New Zealand cicadas in three genera based on multiple independent nuclear introns and mitochondrial DNA: the Kikiha muta complex, the Maoricicada campbelli complex, and the Rhodopsalta species complex. Analyses of molecular data, in combination with song and morphological differences, will improve our understanding of how genetic differences among populations that are geographically isolated to varying degrees may eventually lead to speciation. It will also allow us to test a number of interesting biogeographic hypothesis based on the well-studied paleo-climatological and geological history of New Zealand. In the process we will gain a more complete picture of NZ cicada species diversity (broadly surveyed in the 1960s-70s). The islands of NZ are intriguing because they include a biota composed of both continental and island elements and a range of habitats as diverse as alpine tundra, sand dunes and subtropical to cold temperate rainforest. Our previous work suggests that cicadas arrived in NZ as two colonizing species--one with Australian affinities and one with New Caledonian affinities--about 10 m years ago and subsequently evolved into approximately 40-60 species occupying virtually all habitat types. Our approach of constructing phylogeographic trees for multiple independent nuclear and mitochondrial DNA data sets is necessary because species histories are complicated by such factors as hybridization and random inheritance of polymorphic ancestral genotypes that can only be recognized when independent genes are compared. We have created detailed phylogeographic trees for three NZ cicada species complexes. Each of these species groups has been sampled from many parts of the species range; multiple cryptic species have been discovered. Courtship songs have been recorded for a majority of the individuals sampled. Speciation events have been time calibrated using a variety of relaxed-molecular-clock methods and we have studied diversification rates through time. Publications include:
-
Buckley, T.R., C. Simon, and G.K. Chambers. 2001. Exploring among-site rate variation models in a maximum likelihood framework using empirical data: the effects of model assumptions on estimates of topology, branch lengths, and bootstrap support. Syst. Biol. 50:67-86.
-
Buckley, T.R. C. Simon, H. Shimodaira, and G. K. Chambers. 2001. Evaluating hypotheses on the origin and evolution of the New Zealand alpine cicadas (Maoricicada) using multiple comparison tests of tree topology. Molecular Biology and Evolution 18(2): 223234.
-
Buckley, T.R., C. Simon, and G.K. Chambers. 2001. Phylogeography of the New Zealand cicada Maoricicada campbelli based on mitochondrial DNA sequences: Ancient clades associated with Cenozoic environmental change. Evolution 55:1395-1407.
-
Arensburger, P., T.R. Buckley, C. Simon, M. Moulds and K. E. Holsinger. 2004. Biogeography and phylogeny of the New Zealand Cicada Genera (Hemiptera: Cicadidae) based on nuclear and mitochondrial DNA data. Journal of Biogeography 31: 557569.
-
Arensburger, P., C. Simon, and K. Holsinger. 2004. Evolution and Phylogeny of the New Zealand cicada genus Kikihia Dugdale (Homoptera: Auchenorrhyncha: Cicadidae) with special reference to the origin of the Kermadec and Norfolk Islands species. Journal of Biogeography, 31: 1769-1783.
-
Buckley, T.R., M. Cordeiro, D.C. Marshall, and C. Simon. 2006. Differentiating between hypotheses of lineage sorting and introgression in New Zealand alpine cicadas (Maoricicada Dugdale). Systematic Biology 55:411-425.
-
Marshall, D.C., K. Slon, J.R. Cooley, K.R.B. Hill, and C. Simon. In press. Steady Plio-Pleistocene diversification and a 2-Million-Year sympatry threshold in a New Zealand cicada radiation. Molecular Phylogenetics and Evolution.
-
Hill, K.R.B., C. Simon, D.C. Marshall, and G.K. Chambers. Submitted. Surviving glacial ages within the biotic gap: Phylogeography of the New Zealand cicada Maoricicada campbelli. Minor revisions completed and resubmitted to J. Biogeography.
Possible smaller future projects include phylogeographic studies of species that have not yet been studied in great detail including: the Maoricicada mangu group; the Kikihia cutora group, Kikihia murihikua (vs. K. angusta and K. rosea/balaena); and K. tasmani and relatives (note that the we are not certain whether we have indeed found the true mtDNA of K. tasmani or K. murihikua.
|
Gene Flow and Contact zones in New Zealand cicada species
Beth Wade1, Chris Simon1, 2, Dave Marshall1, Kathy Hill1, and John Cooley1,3
1The University of Connecticut
2Victoria University, Wellington
3Yale University
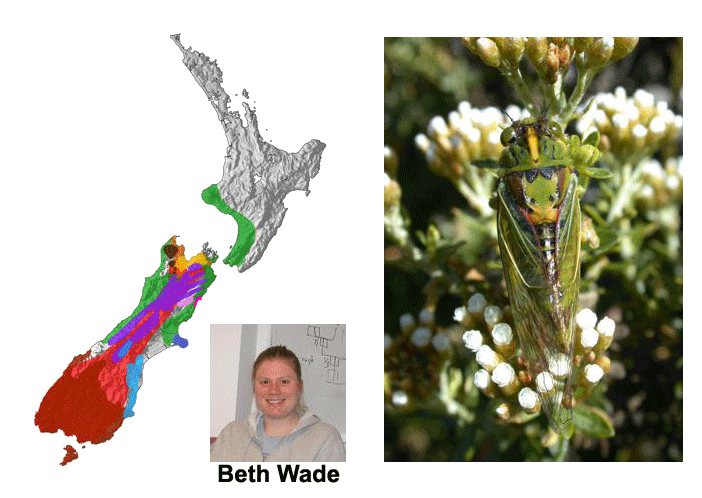 This project is a follow-up to a collaboration between the Simon Lab and the Chambers lab in NZ. That project was supported by US NSF and NZ Marsden grants and ran from 2000 to 2004. What Factors Determine the Level of Gene Flow Between Natural Populations of Organisms? The cessation of gene flow between two formerly interbreeding populations is thought to play a key role in speciation. Filters to gene flow can include geographic, temporal, and/or behavioral components. There has been much debate in the literature about the nature of species boundaries. Recent work suggests that gene exchange among species may be more common than previously suspected and that the idea of reproductively isolated biological species may in many cases be an oversimplification. We propose to investigate the nature of species boundaries by studying the interactions of adjacent and/or overlapping species of varying degrees of relatedness. As new species form and populations diverge from each other (due to lack of gene flow, natural selection and/or chance), the sexual signals that help animals recognize their mates also diverge. In many cases, rapidly changing environments, like those that have characterized the paleo- and recent history of NZ, result in secondary contact of once isolated populations. With secondary contact, formerly isolated populations can: 1) remain distinct, reproductively-isolated species, becoming even more distinct where they overlap; 2) interbreed and become one indistinguishable group; 3) interbreed, form a persistent hybrid zone, but remain distinct outside this zone due to selection against introgression of foreign genes; or 4) as J. Mallet has suggested for Heliconius butterflies of the Amazon, interbreed, exchange genes extensively but still remain diagnosable species. In each of the possible scenarios, sexual signals play a key role in determining the outcome. We propose to use NZ cicadas to test the fundamental evolutionary predictions that: 1) the sexual signals (songs) of different species will, at intermediate genetic distances, differ more where these populations overlap than where they do not; 2) there is a threshold below which sexual signals and genetic distances will be too similar to trigger this evolutionary response and populations will hybridize; and 3) there is a point beyond which sexual signals and genetic distances are so different that no response occurs. Where hybridization is found to occur, we will determine the extent of gene flow using microsatellite markers. Dugdale, Fleming, and Lane detailed many localities at which species or subspecies of NZ cicadas come into contact; we have found additional contact zones in our field surveys of the last 15 years. Our study would be one of the first to analyze inter- and intraspecific variation in acoustic sexual signals of a large, ecologically diverse animal taxon in an observational, experimental, and evolutionary context. The new project would take advantage of results produced by our NZ phylogeography project including a very comprehensive phylogeny of the genus Kikihia, recordings of all Kikihia songs, large collections of cicadas from contact zones paired with collections of presumed parental types away from contact zones. There are more than 19 locations in New Zealand where hybrids have been discovered between various species pairs. In addition, the Marsden project developed primers for and characterized three microsatellite loci that can be amplified in most Kikihia species. Data has been collected for contact zone versus parental populations for one of these loci.
This project is a follow-up to a collaboration between the Simon Lab and the Chambers lab in NZ. That project was supported by US NSF and NZ Marsden grants and ran from 2000 to 2004. What Factors Determine the Level of Gene Flow Between Natural Populations of Organisms? The cessation of gene flow between two formerly interbreeding populations is thought to play a key role in speciation. Filters to gene flow can include geographic, temporal, and/or behavioral components. There has been much debate in the literature about the nature of species boundaries. Recent work suggests that gene exchange among species may be more common than previously suspected and that the idea of reproductively isolated biological species may in many cases be an oversimplification. We propose to investigate the nature of species boundaries by studying the interactions of adjacent and/or overlapping species of varying degrees of relatedness. As new species form and populations diverge from each other (due to lack of gene flow, natural selection and/or chance), the sexual signals that help animals recognize their mates also diverge. In many cases, rapidly changing environments, like those that have characterized the paleo- and recent history of NZ, result in secondary contact of once isolated populations. With secondary contact, formerly isolated populations can: 1) remain distinct, reproductively-isolated species, becoming even more distinct where they overlap; 2) interbreed and become one indistinguishable group; 3) interbreed, form a persistent hybrid zone, but remain distinct outside this zone due to selection against introgression of foreign genes; or 4) as J. Mallet has suggested for Heliconius butterflies of the Amazon, interbreed, exchange genes extensively but still remain diagnosable species. In each of the possible scenarios, sexual signals play a key role in determining the outcome. We propose to use NZ cicadas to test the fundamental evolutionary predictions that: 1) the sexual signals (songs) of different species will, at intermediate genetic distances, differ more where these populations overlap than where they do not; 2) there is a threshold below which sexual signals and genetic distances will be too similar to trigger this evolutionary response and populations will hybridize; and 3) there is a point beyond which sexual signals and genetic distances are so different that no response occurs. Where hybridization is found to occur, we will determine the extent of gene flow using microsatellite markers. Dugdale, Fleming, and Lane detailed many localities at which species or subspecies of NZ cicadas come into contact; we have found additional contact zones in our field surveys of the last 15 years. Our study would be one of the first to analyze inter- and intraspecific variation in acoustic sexual signals of a large, ecologically diverse animal taxon in an observational, experimental, and evolutionary context. The new project would take advantage of results produced by our NZ phylogeography project including a very comprehensive phylogeny of the genus Kikihia, recordings of all Kikihia songs, large collections of cicadas from contact zones paired with collections of presumed parental types away from contact zones. There are more than 19 locations in New Zealand where hybrids have been discovered between various species pairs. In addition, the Marsden project developed primers for and characterized three microsatellite loci that can be amplified in most Kikihia species. Data has been collected for contact zone versus parental populations for one of these loci.
Possible smaller project related to the above: To extract DNA from known hybrid populations and to clone and sequence multiple sequences from single individuals to look for obvious paternal leakage of mtDNA, and other non-standard copies of mtDNA (as in item 1 above). This is a project that would be guaranteed to produce interesting results.
|
Bar coding, taxonomy, and New Zealand endemics
Pete Ritchie2, David Chappell2, Megan Ribak1, and Chris Simon1,2
1The University of Connecticut
2Victoria University, Wellington
This is a proposed project that would be a joint barcoding efforts including skinks (Ritchie, Chappell) and cicadas (Simon). The following description comes from an abstract written by Pete Ritchie and David Chapple with cicada information added by C. Simon. The correct identification of species underpins all biological studies. Although the biological sciences are reliant on the ability to efficiently distinguish meaningful groups in nature, there is a growing concern that traditional taxonomy is too cumbersome to achieve this goal. Recently the International Consortium for the Barcode of Life attempted to revolutionize the traditional biological classification system by suggesting that we should classify all living species by their DNA barcode. This technique uses a short (650+ bp) standardized stretch of DNA sequence from the mitochondrial COI gene to genetically identify ('barcode') known species and to find new ones. Data can be stored and retrieved electronically. However, the methodology needs extensive testing and verification by reference to other DNA markers. We propose to DNA barcode all of the New Zealand skinks and all the NZ cicadas, the first such endeavour for two species Families in New Zealand, and test the DNA barcoding method with different analysis methods and a range of comparable genetic markers. This project has the advantage that a multigene phylogeny for all NZ skinks and cicadas is nearly complete, the samples are in hand, and we lack only the bar code sequences. This new bardcoding scheme has generated tremendous debate in the scientific community regarding the nature of systematics and science in general.
This project would involve sequencing the barcode segment of the COI gene for all NZ cicadas in the Kikihia, Maoricicada, and Rhodopsalta lineages. By chance, the bar coders chose the half of the COI gene that we have not yet sequenced. Once the sequences are collected, phylogenetic trees can be constructed using the bar code piece alone and compared to trees constructed from much larger mtDNA data sets. Another good undergrad project would be to look at COI genes from all the cicada species we have sequenced to document the amino acid conservation (should be about 70%) and to compare it to other invertebrates as in Lunt, D.H., Zhang, D., Syzmura, J. & Hewitt, G.M. (1996) The insect cytochrome oxidase I gene: evolutionary patterns and conserved primers for phylogenetic studies. Insect Bichemistry and Molecular Biology 5, 153-165.
Undergraduate researcher Megan Ribak is assessing the efficacy of the bar-coding region of the mitochondrial COI gene for constructing a phylogeny of the genus Kikihia and for identifying known species. She will compare her phylogenetic results to those from a nuclear and mitochondrial DNA (mtDNA) data set that have already been collected for this genus by Postdoc David Marshall as part of our current NSF award. In addition, all Kikihia species have been characterized by their song; a comparison of mtDNA type and song can help recognize hybrids that could be a challenge for bar coding.
|
Biodiversity, Taxonomy, Systematics and Evolution of Cicadas Worldwide
Simon Lab (University of Connecticut), Dietrich Lab (University of Illinois), Cryan Lab (NY State Museum of Natural History)
 This is a PEET partnership. The Simon lab group is handing the cicada part of the grant. We collaborate with cicada experts worldwide, especially Max Moulds in Australia and Allen Sanborn in Miami. Young June Lee, a postdoc from Korea, and Geert Goemans, a UCONN Ph.D. student will be participating in the project along with other Simon Lab members such as Technician Kathy Hill, postdoc Dave Marshall, and Research Associate John Cooley.
Innovative aspects of the project include its coordinated approach to training, involving several experts with complementary expertise; the use of advanced tools for the efficient synthesis of taxonomic information and production of improved classifications and identification tools in electronic and traditional formats; and the applicability of the anticipated phylogenetic results to basic conceptual issues in systematics and evolution. Click here for more information on this project.
This is a PEET partnership. The Simon lab group is handing the cicada part of the grant. We collaborate with cicada experts worldwide, especially Max Moulds in Australia and Allen Sanborn in Miami. Young June Lee, a postdoc from Korea, and Geert Goemans, a UCONN Ph.D. student will be participating in the project along with other Simon Lab members such as Technician Kathy Hill, postdoc Dave Marshall, and Research Associate John Cooley.
Innovative aspects of the project include its coordinated approach to training, involving several experts with complementary expertise; the use of advanced tools for the efficient synthesis of taxonomic information and production of improved classifications and identification tools in electronic and traditional formats; and the applicability of the anticipated phylogenetic results to basic conceptual issues in systematics and evolution. Click here for more information on this project.
Possible undergrad projects related to this PEET project: To choose a small genus of cicadas and use it to learn the tools of taxonomy including: collecting specimens in the field, identifying and classifying cicadas, curating specimens, DNA extraction and amplification, molecular phylogenetics, micro- and macro- photography, 2- and 3-dimensional digital imaging, key construction, and web dissemination of information.
|
Systematics and Biogeography of Cicadettini world wide
David Marshall1, Kathy Hill1, Chris Owen1, Dan Vanderpool1, Max Moulds4, Thomas Buckley3, Peter Ritchie2, John Cooley1,5, and Chris Simon1,2
Matija Gogala, Tomi Trilar, Martin Villet, Jason Cryan, Chris Dietrich, Allen Sanborn, Zhongren Li, Arnold de Boer, Marieke Schouten, Jerome Sueur, Stephane Puissant, Jian Hong Chen
1The University of Connecticut
2Victoria University, Wellington
3Landcare, Auckland
4The Australian Museum, Sydney
5Yale University
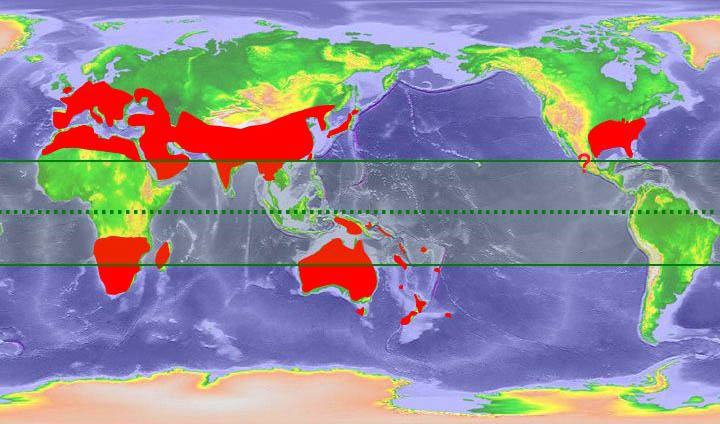 This proposal is a collaboration that involves researchers in the US, China, Australia, NZ, Slovenia, France, the Netherlands, and South Africa, with the goal of reconstructing the evolutionary history of the worldwide cicada tribe Cicadettini. Preliminary fieldwork suggests that a rich diversity of species remains to be discovered. We aim to provide a thorough study of how a large taxonomic group diversifies and spreads worldwide. Since the present-day center of diversity of Cicadettini is Australia, it will be useful to learn more about the timing of Australian species radiations and the relationships of these events to paleoclimate and landscape changes. Our specific goals are to: 1) complete a comprehensive phylogeny of Cicadettini adding at least one species from each of the 20+ described but uncollected genera and increasing the number of taxa studied from 80 to 300; 2) test the monophyly of Cicadettini using outgroup taxa from four related tribes; 3) assess the monophyly of cicadettine genera and explore in detail all remaining Australian taxa formerly placed in Cicadetta; 4) construct phylogenetic trees for the largest, most widespread Australian cicadettine genera to establish calibrations for molecular dating; 5) use relaxed-clock dating and lineage-through-time plots to relate clade formation to Australian and worldwide climate shifts; and 6) expand our website Cicada Central. Cicadas make excellent model systems for studies of biogeographic patterns because their acoustic sexual signals make them easy to collect and identify and their sedentary life history results in clear biogeographic patterning. Cicadas occupy a broad range of habitats and are distributed on all continents except Antarctica. They are known from Mesozoic and Cenozoic fossils. Cicadettini represent approximately 1/3 of cicada diversity world-wide. Previous work in each of our labs suggests that this project will be successful. We have a strong team of researchers from three countries with complementary expertise in molecular, morphological, and acoustic analyses. The team includes senior researchers, early- career scientists, and graduate and undergraduate students who will all receive training as part of this project. The longer-term goals of our group are to describe the remaining NZ and Australian cicada species, to construct a phylogenetic tree for the family Cicadidae (3000 species worldwide) that will contribute to the international effort to construct a Tree of Life. Specimens collected as part of the present proposal will include all cicada lineages (for future projects) and other homopteran relatives (for colleagues). Each collecting trip we have made in our preliminary work has turned up 50 to 80 percent undescribed species.
This proposal is a collaboration that involves researchers in the US, China, Australia, NZ, Slovenia, France, the Netherlands, and South Africa, with the goal of reconstructing the evolutionary history of the worldwide cicada tribe Cicadettini. Preliminary fieldwork suggests that a rich diversity of species remains to be discovered. We aim to provide a thorough study of how a large taxonomic group diversifies and spreads worldwide. Since the present-day center of diversity of Cicadettini is Australia, it will be useful to learn more about the timing of Australian species radiations and the relationships of these events to paleoclimate and landscape changes. Our specific goals are to: 1) complete a comprehensive phylogeny of Cicadettini adding at least one species from each of the 20+ described but uncollected genera and increasing the number of taxa studied from 80 to 300; 2) test the monophyly of Cicadettini using outgroup taxa from four related tribes; 3) assess the monophyly of cicadettine genera and explore in detail all remaining Australian taxa formerly placed in Cicadetta; 4) construct phylogenetic trees for the largest, most widespread Australian cicadettine genera to establish calibrations for molecular dating; 5) use relaxed-clock dating and lineage-through-time plots to relate clade formation to Australian and worldwide climate shifts; and 6) expand our website Cicada Central. Cicadas make excellent model systems for studies of biogeographic patterns because their acoustic sexual signals make them easy to collect and identify and their sedentary life history results in clear biogeographic patterning. Cicadas occupy a broad range of habitats and are distributed on all continents except Antarctica. They are known from Mesozoic and Cenozoic fossils. Cicadettini represent approximately 1/3 of cicada diversity world-wide. Previous work in each of our labs suggests that this project will be successful. We have a strong team of researchers from three countries with complementary expertise in molecular, morphological, and acoustic analyses. The team includes senior researchers, early- career scientists, and graduate and undergraduate students who will all receive training as part of this project. The longer-term goals of our group are to describe the remaining NZ and Australian cicada species, to construct a phylogenetic tree for the family Cicadidae (3000 species worldwide) that will contribute to the international effort to construct a Tree of Life. Specimens collected as part of the present proposal will include all cicada lineages (for future projects) and other homopteran relatives (for colleagues). Each collecting trip we have made in our preliminary work has turned up 50 to 80 percent undescribed species.
Possible undergraduate projects include choosing a genus in the tribe Cicadettini and learning to extract, amplify, and sequence DNA data and then to analyze these data phylogenetically and biogeographically.
|
 Undergraduate researcher Rachel Krauss studied the phylogenetics, phylogeography and biogeography of the Australian cicada genus Gudanga whose nine putative species are distributed across arid Australia and are related to the Australian genus Pauropsalta (Marshall, Hill, Vanderpool, and Simon, unpubl). Gudanga species are interesting because they have very subtle (or no) song differences between some of the named species while all other cicada species are distinguished by species-specific calling songs. Gudanga species are mostly described from hind-wing and abdominal color patterns and some of these are recorded as occurring sympatrically. Rachel explored genetic variation within and among species and created a maximum likelihood phylogenetic tree of taxa based on mitochondrial COI and COII gene segments plus nuclear Elongation-Factor-1-alpha sequence. She mapped her tree onto the landscape of Australia and discovered three geographically coherent, cryptic, potential new species. She dissected, cleared, photographed (using Automontage) and drew the genitalia. She described the color patterns and measured each of the putative species identified by the DNA data. Rachel presented her work as a poster at the UCONN campuswide "Frontiers in Undergraduate Research Colloquium" (see photo above).
Undergraduate researcher Rachel Krauss studied the phylogenetics, phylogeography and biogeography of the Australian cicada genus Gudanga whose nine putative species are distributed across arid Australia and are related to the Australian genus Pauropsalta (Marshall, Hill, Vanderpool, and Simon, unpubl). Gudanga species are interesting because they have very subtle (or no) song differences between some of the named species while all other cicada species are distinguished by species-specific calling songs. Gudanga species are mostly described from hind-wing and abdominal color patterns and some of these are recorded as occurring sympatrically. Rachel explored genetic variation within and among species and created a maximum likelihood phylogenetic tree of taxa based on mitochondrial COI and COII gene segments plus nuclear Elongation-Factor-1-alpha sequence. She mapped her tree onto the landscape of Australia and discovered three geographically coherent, cryptic, potential new species. She dissected, cleared, photographed (using Automontage) and drew the genitalia. She described the color patterns and measured each of the putative species identified by the DNA data. Rachel presented her work as a poster at the UCONN campuswide "Frontiers in Undergraduate Research Colloquium" (see photo above).
|
Dispersal, Vicariance, and the geologic history of New Zealand
Peter Ritchie2, George Gibbs2, Chris Simon1,2, Thomas Buckley3, Ian Henderson4, Karl Kjer5, and Kjell Arne Johanson6
1The University of Connecticut
2Victoria University, Wellington
3Landcare, Auckland
4Massey University
5Rutgers University
6Swedish Museum of Natural History
|
This is a collaborative proposal addressing the following question: Is New Zealand a continental fragment or an oceanic island? We will submit a grant to the NZ Marsden fund to support this research. The relative influences of vicariance and dispersal on the distribution of biodiversity have been a central debate in historical biogeography. New Zealand has become a focal point in this debate owing to its unique geological history and present geographical isolation. Two non-mutually exclusive hypotheses have been proposed to explain the origin of the New Zealand biota: Hypothesis 1: New Zealand has the biota of a Gondwanan continental fragment that retained many original lineages since its vicariant separation. Hypothesis 2: New Zealand has the biota of an oceanic island that was replenished by transoceanic dispersal following extinction during the Oligocene drowning event. Waters and Craw make the case that all current molecular dating studies are either ambiguous or support hypothesis 2. However, this lack of direct evidence for the Gondwanan origin of the New Zealand biota is in part due to a focus on organisms with high dispersal ability, a shortage of robust molecular dating analyses, and no clear testable framework. Therefore, the central question on the origins of New Zealands unique biota remains unresolved. The shape of a community phylogeny: A new approach to an old problem. We propose to develop a new theoretical framework - the concept of testable community phylogenies by adapting coalescence theory, and applying it to phylogenies of the New Zealand freshwater invertebrate community members to critically test the above two hypotheses. We will focus on the freshwater invertebrate community (specifically caddisflies) they are poor dispersers that are highly endemic and restricted to their non-marine aquatic home. In addition, several published studies of stream invertebrate species have shown characteristically Gondwanan sister-group relationships between South America (SA) and Australia (AUS), rather than AUS and New Zealand (NZ), the former pattern being consistent with vicariance. The obligate habitat of freshwater invertebrates defines an ecological community and this coherence will allow us to reconstruct a community phylogeny by summing the phylogenies of component taxa. Our new approach will utilise the general shape of this community phylogeny and dates of divergence to infer the past processes that have affected the present day community structure. To create a testable framework we will adapt coalescence theory developed to study the processes that shape gene genealogies and establish an explicit set of possible community phylogenies that would arise under different conditions. The forces that affect species diversity in a community (which are analogues to the forces that affect genetic diversity a population) that we will quantify are summarised as: i) speciation (mutation); ii) random ecological drift; iii) migration of species among communities; and iv) selection of one species over another. The two hypotheses above make the following predictions: Hypothesis 1 (Vicariant origin of the community): 1) Speciation: a ((SA+AUS)NZ) phylogenetic pattern, with divergence dates matching tectonic events. 2) Random ecological drift: gradual extinctions since Gondwana, increased rate during Oligocene and climate change events. 3) Migration: sporadic between NZ and AUS since break up of Gondwana.
Hypothesis 2 (Community derived from dispersal): 4) Speciation: Absence of ((SA+AUS)NZ), with ((NZ+AUS)SA) pattern predominating: no NZ+AUS divergence dates older than Oligocene drowning. 5) Random ecological drift: Complete extinction during Oligocene and increased rat during climate change events. 6) Migration: No evidence before Oligocene. We propose to sample representative tricopteran taxa from the cool, clear streams of Chile, New Zealand and Australia. We will use protein-coding nuclear and mitochondrial genes to reconstruct the phylogeny of each focal taxon using standard phylogenetic methods. Dates of divergences will be estimated using previously determined rates of evolution and biogeographic calibrations, external to the biogeographic questions that we will test. The proposed research will not only provide information about the origins of New Zealand freshwater fauna and the dynamics of communities over long periods of time, but it will facilitate the development of a new synthetic analytical approach for examining the evolution and phylogeny of community assemblages. |
Long-term goals of the Simon Lab
|
The longer-term goals of our lab groups are to continue to address a broad range of important biological questions relating to: the origin, evolution, and spread of biological diversity; speciation; the evolution of life histories; the evolution of courtship song and its role in speciation; the estimation of times of divergence in evolutionary trees; and application of information on molecular evolutionary processes to phylogenetic tree-building. Our phylogeographic studies of NZ cicadas take advantage of an already established international collaboration with a large amount of preliminary data and a well-developed knowledge of the natural history of NZ. Our research group is broader than the personnel named in any particular proposal and includes investigators in Australia, NZ, USA, Africa, Asia and Europe. The UCONN lab includes three postdoctoral associates, five grad students, and three undergraduate researchers; the NZ lab has a number of professional researchers addressing related questions. Trainees learn skills essential in population biology, e.g., specimen collection; database construction; collection and analysis of genetic and behavioral data; and written, oral, and web-based data dissemination. Part of the team is involved in a world-wide study of various genera and tribes of cicadas and collaborating with a larger team whose goal is to describe the evolution of the paraneopteran insects (thrips, lice, plant-sucking bugs and true bugs) to contribute to the global effort to track the evolution of all life.
|
|

 Animal mitochondrial DNA (mtDNA) is generally assumed to be maternally inherited. Is this assumption correct? In the last 20 years, several instances have been discovered in which animal mtDNA is transmitted through patrilines, a phenomenon termed "paternal leakage". It challenges some of the assumptions involved in using mtDNA as a molecular or forensic marker and it can complicate phylogenetic reconstruction and molecular dating. We have demonstrated paternal leakage in experimental hybrid crosses of periodical cicadas, and we have taken advantage of the phenomenon of paternal leakage to demonstrate that male periodical cicadas can gain from mating an already-mated female. These results provide important clues about why male periodical cicadas are so persistent in approaching and courting even unreceptive females. Read more about this project here.
Animal mitochondrial DNA (mtDNA) is generally assumed to be maternally inherited. Is this assumption correct? In the last 20 years, several instances have been discovered in which animal mtDNA is transmitted through patrilines, a phenomenon termed "paternal leakage". It challenges some of the assumptions involved in using mtDNA as a molecular or forensic marker and it can complicate phylogenetic reconstruction and molecular dating. We have demonstrated paternal leakage in experimental hybrid crosses of periodical cicadas, and we have taken advantage of the phenomenon of paternal leakage to demonstrate that male periodical cicadas can gain from mating an already-mated female. These results provide important clues about why male periodical cicadas are so persistent in approaching and courting even unreceptive females. Read more about this project here.


 This project, funded by the U.S. National Science Foundation, addresses questions of how past changes in climate and land forms affect speciation. New combinations of phylogenetic and phylogeographic techniques using multiple gene loci are becoming key tools in the description of genetic diversity within species. These approaches can test detailed hypotheses of population and species origin, range changes, and extinction. We are conducting a fine-scale phylogeographic study of three widely co-distributed species complexes of New Zealand cicadas in three genera based on multiple independent nuclear introns and mitochondrial DNA: the Kikiha muta complex, the Maoricicada campbelli complex, and the Rhodopsalta species complex. Analyses of molecular data, in combination with song and morphological differences, will improve our understanding of how genetic differences among populations that are geographically isolated to varying degrees may eventually lead to speciation. It will also allow us to test a number of interesting biogeographic hypothesis based on the well-studied paleo-climatological and geological history of New Zealand. In the process we will gain a more complete picture of NZ cicada species diversity (broadly surveyed in the 1960s-70s). The islands of NZ are intriguing because they include a biota composed of both continental and island elements and a range of habitats as diverse as alpine tundra, sand dunes and subtropical to cold temperate rainforest. Our previous work suggests that cicadas arrived in NZ as two colonizing species--one with Australian affinities and one with New Caledonian affinities--about 10 m years ago and subsequently evolved into approximately 40-60 species occupying virtually all habitat types. Our approach of constructing phylogeographic trees for multiple independent nuclear and mitochondrial DNA data sets is necessary because species histories are complicated by such factors as hybridization and random inheritance of polymorphic ancestral genotypes that can only be recognized when independent genes are compared. We have created detailed phylogeographic trees for three NZ cicada species complexes. Each of these species groups has been sampled from many parts of the species range; multiple cryptic species have been discovered. Courtship songs have been recorded for a majority of the individuals sampled. Speciation events have been time calibrated using a variety of relaxed-molecular-clock methods and we have studied diversification rates through time. Publications include:
This project, funded by the U.S. National Science Foundation, addresses questions of how past changes in climate and land forms affect speciation. New combinations of phylogenetic and phylogeographic techniques using multiple gene loci are becoming key tools in the description of genetic diversity within species. These approaches can test detailed hypotheses of population and species origin, range changes, and extinction. We are conducting a fine-scale phylogeographic study of three widely co-distributed species complexes of New Zealand cicadas in three genera based on multiple independent nuclear introns and mitochondrial DNA: the Kikiha muta complex, the Maoricicada campbelli complex, and the Rhodopsalta species complex. Analyses of molecular data, in combination with song and morphological differences, will improve our understanding of how genetic differences among populations that are geographically isolated to varying degrees may eventually lead to speciation. It will also allow us to test a number of interesting biogeographic hypothesis based on the well-studied paleo-climatological and geological history of New Zealand. In the process we will gain a more complete picture of NZ cicada species diversity (broadly surveyed in the 1960s-70s). The islands of NZ are intriguing because they include a biota composed of both continental and island elements and a range of habitats as diverse as alpine tundra, sand dunes and subtropical to cold temperate rainforest. Our previous work suggests that cicadas arrived in NZ as two colonizing species--one with Australian affinities and one with New Caledonian affinities--about 10 m years ago and subsequently evolved into approximately 40-60 species occupying virtually all habitat types. Our approach of constructing phylogeographic trees for multiple independent nuclear and mitochondrial DNA data sets is necessary because species histories are complicated by such factors as hybridization and random inheritance of polymorphic ancestral genotypes that can only be recognized when independent genes are compared. We have created detailed phylogeographic trees for three NZ cicada species complexes. Each of these species groups has been sampled from many parts of the species range; multiple cryptic species have been discovered. Courtship songs have been recorded for a majority of the individuals sampled. Speciation events have been time calibrated using a variety of relaxed-molecular-clock methods and we have studied diversification rates through time. Publications include:
 This project is a follow-up to a collaboration between the Simon Lab and the Chambers lab in NZ. That project was supported by US NSF and NZ Marsden grants and ran from 2000 to 2004. What Factors Determine the Level of Gene Flow Between Natural Populations of Organisms? The cessation of gene flow between two formerly interbreeding populations is thought to play a key role in speciation. Filters to gene flow can include geographic, temporal, and/or behavioral components. There has been much debate in the literature about the nature of species boundaries. Recent work suggests that gene exchange among species may be more common than previously suspected and that the idea of reproductively isolated biological species may in many cases be an oversimplification. We propose to investigate the nature of species boundaries by studying the interactions of adjacent and/or overlapping species of varying degrees of relatedness. As new species form and populations diverge from each other (due to lack of gene flow, natural selection and/or chance), the sexual signals that help animals recognize their mates also diverge. In many cases, rapidly changing environments, like those that have characterized the paleo- and recent history of NZ, result in secondary contact of once isolated populations. With secondary contact, formerly isolated populations can: 1) remain distinct, reproductively-isolated species, becoming even more distinct where they overlap; 2) interbreed and become one indistinguishable group; 3) interbreed, form a persistent hybrid zone, but remain distinct outside this zone due to selection against introgression of foreign genes; or 4) as J. Mallet has suggested for Heliconius butterflies of the Amazon, interbreed, exchange genes extensively but still remain diagnosable species. In each of the possible scenarios, sexual signals play a key role in determining the outcome. We propose to use NZ cicadas to test the fundamental evolutionary predictions that: 1) the sexual signals (songs) of different species will, at intermediate genetic distances, differ more where these populations overlap than where they do not; 2) there is a threshold below which sexual signals and genetic distances will be too similar to trigger this evolutionary response and populations will hybridize; and 3) there is a point beyond which sexual signals and genetic distances are so different that no response occurs. Where hybridization is found to occur, we will determine the extent of gene flow using microsatellite markers. Dugdale, Fleming, and Lane detailed many localities at which species or subspecies of NZ cicadas come into contact; we have found additional contact zones in our field surveys of the last 15 years. Our study would be one of the first to analyze inter- and intraspecific variation in acoustic sexual signals of a large, ecologically diverse animal taxon in an observational, experimental, and evolutionary context. The new project would take advantage of results produced by our NZ phylogeography project including a very comprehensive phylogeny of the genus Kikihia, recordings of all Kikihia songs, large collections of cicadas from contact zones paired with collections of presumed parental types away from contact zones. There are more than 19 locations in New Zealand where hybrids have been discovered between various species pairs. In addition, the Marsden project developed primers for and characterized three microsatellite loci that can be amplified in most Kikihia species. Data has been collected for contact zone versus parental populations for one of these loci.
This project is a follow-up to a collaboration between the Simon Lab and the Chambers lab in NZ. That project was supported by US NSF and NZ Marsden grants and ran from 2000 to 2004. What Factors Determine the Level of Gene Flow Between Natural Populations of Organisms? The cessation of gene flow between two formerly interbreeding populations is thought to play a key role in speciation. Filters to gene flow can include geographic, temporal, and/or behavioral components. There has been much debate in the literature about the nature of species boundaries. Recent work suggests that gene exchange among species may be more common than previously suspected and that the idea of reproductively isolated biological species may in many cases be an oversimplification. We propose to investigate the nature of species boundaries by studying the interactions of adjacent and/or overlapping species of varying degrees of relatedness. As new species form and populations diverge from each other (due to lack of gene flow, natural selection and/or chance), the sexual signals that help animals recognize their mates also diverge. In many cases, rapidly changing environments, like those that have characterized the paleo- and recent history of NZ, result in secondary contact of once isolated populations. With secondary contact, formerly isolated populations can: 1) remain distinct, reproductively-isolated species, becoming even more distinct where they overlap; 2) interbreed and become one indistinguishable group; 3) interbreed, form a persistent hybrid zone, but remain distinct outside this zone due to selection against introgression of foreign genes; or 4) as J. Mallet has suggested for Heliconius butterflies of the Amazon, interbreed, exchange genes extensively but still remain diagnosable species. In each of the possible scenarios, sexual signals play a key role in determining the outcome. We propose to use NZ cicadas to test the fundamental evolutionary predictions that: 1) the sexual signals (songs) of different species will, at intermediate genetic distances, differ more where these populations overlap than where they do not; 2) there is a threshold below which sexual signals and genetic distances will be too similar to trigger this evolutionary response and populations will hybridize; and 3) there is a point beyond which sexual signals and genetic distances are so different that no response occurs. Where hybridization is found to occur, we will determine the extent of gene flow using microsatellite markers. Dugdale, Fleming, and Lane detailed many localities at which species or subspecies of NZ cicadas come into contact; we have found additional contact zones in our field surveys of the last 15 years. Our study would be one of the first to analyze inter- and intraspecific variation in acoustic sexual signals of a large, ecologically diverse animal taxon in an observational, experimental, and evolutionary context. The new project would take advantage of results produced by our NZ phylogeography project including a very comprehensive phylogeny of the genus Kikihia, recordings of all Kikihia songs, large collections of cicadas from contact zones paired with collections of presumed parental types away from contact zones. There are more than 19 locations in New Zealand where hybrids have been discovered between various species pairs. In addition, the Marsden project developed primers for and characterized three microsatellite loci that can be amplified in most Kikihia species. Data has been collected for contact zone versus parental populations for one of these loci.
 This is a PEET partnership. The Simon lab group is handing the cicada part of the grant. We collaborate with cicada experts worldwide, especially Max Moulds in Australia and Allen Sanborn in Miami. Young June Lee, a postdoc from Korea, and Geert Goemans, a UCONN Ph.D. student will be participating in the project along with other Simon Lab members such as Technician Kathy Hill, postdoc Dave Marshall, and Research Associate John Cooley.
Innovative aspects of the project include its coordinated approach to training, involving several experts with complementary expertise; the use of advanced tools for the efficient synthesis of taxonomic information and production of improved classifications and identification tools in electronic and traditional formats; and the applicability of the anticipated phylogenetic results to basic conceptual issues in systematics and evolution. Click here for more information on this project.
This is a PEET partnership. The Simon lab group is handing the cicada part of the grant. We collaborate with cicada experts worldwide, especially Max Moulds in Australia and Allen Sanborn in Miami. Young June Lee, a postdoc from Korea, and Geert Goemans, a UCONN Ph.D. student will be participating in the project along with other Simon Lab members such as Technician Kathy Hill, postdoc Dave Marshall, and Research Associate John Cooley.
Innovative aspects of the project include its coordinated approach to training, involving several experts with complementary expertise; the use of advanced tools for the efficient synthesis of taxonomic information and production of improved classifications and identification tools in electronic and traditional formats; and the applicability of the anticipated phylogenetic results to basic conceptual issues in systematics and evolution. Click here for more information on this project.
 This proposal is a collaboration that involves researchers in the US, China, Australia, NZ, Slovenia, France, the Netherlands, and South Africa, with the goal of reconstructing the evolutionary history of the worldwide cicada tribe Cicadettini. Preliminary fieldwork suggests that a rich diversity of species remains to be discovered. We aim to provide a thorough study of how a large taxonomic group diversifies and spreads worldwide. Since the present-day center of diversity of Cicadettini is Australia, it will be useful to learn more about the timing of Australian species radiations and the relationships of these events to paleoclimate and landscape changes. Our specific goals are to: 1) complete a comprehensive phylogeny of Cicadettini adding at least one species from each of the 20+ described but uncollected genera and increasing the number of taxa studied from 80 to 300; 2) test the monophyly of Cicadettini using outgroup taxa from four related tribes; 3) assess the monophyly of cicadettine genera and explore in detail all remaining Australian taxa formerly placed in Cicadetta; 4) construct phylogenetic trees for the largest, most widespread Australian cicadettine genera to establish calibrations for molecular dating; 5) use relaxed-clock dating and lineage-through-time plots to relate clade formation to Australian and worldwide climate shifts; and 6) expand our website Cicada Central. Cicadas make excellent model systems for studies of biogeographic patterns because their acoustic sexual signals make them easy to collect and identify and their sedentary life history results in clear biogeographic patterning. Cicadas occupy a broad range of habitats and are distributed on all continents except Antarctica. They are known from Mesozoic and Cenozoic fossils. Cicadettini represent approximately 1/3 of cicada diversity world-wide. Previous work in each of our labs suggests that this project will be successful. We have a strong team of researchers from three countries with complementary expertise in molecular, morphological, and acoustic analyses. The team includes senior researchers, early- career scientists, and graduate and undergraduate students who will all receive training as part of this project. The longer-term goals of our group are to describe the remaining NZ and Australian cicada species, to construct a phylogenetic tree for the family Cicadidae (3000 species worldwide) that will contribute to the international effort to construct a Tree of Life. Specimens collected as part of the present proposal will include all cicada lineages (for future projects) and other homopteran relatives (for colleagues). Each collecting trip we have made in our preliminary work has turned up 50 to 80 percent undescribed species.
This proposal is a collaboration that involves researchers in the US, China, Australia, NZ, Slovenia, France, the Netherlands, and South Africa, with the goal of reconstructing the evolutionary history of the worldwide cicada tribe Cicadettini. Preliminary fieldwork suggests that a rich diversity of species remains to be discovered. We aim to provide a thorough study of how a large taxonomic group diversifies and spreads worldwide. Since the present-day center of diversity of Cicadettini is Australia, it will be useful to learn more about the timing of Australian species radiations and the relationships of these events to paleoclimate and landscape changes. Our specific goals are to: 1) complete a comprehensive phylogeny of Cicadettini adding at least one species from each of the 20+ described but uncollected genera and increasing the number of taxa studied from 80 to 300; 2) test the monophyly of Cicadettini using outgroup taxa from four related tribes; 3) assess the monophyly of cicadettine genera and explore in detail all remaining Australian taxa formerly placed in Cicadetta; 4) construct phylogenetic trees for the largest, most widespread Australian cicadettine genera to establish calibrations for molecular dating; 5) use relaxed-clock dating and lineage-through-time plots to relate clade formation to Australian and worldwide climate shifts; and 6) expand our website Cicada Central. Cicadas make excellent model systems for studies of biogeographic patterns because their acoustic sexual signals make them easy to collect and identify and their sedentary life history results in clear biogeographic patterning. Cicadas occupy a broad range of habitats and are distributed on all continents except Antarctica. They are known from Mesozoic and Cenozoic fossils. Cicadettini represent approximately 1/3 of cicada diversity world-wide. Previous work in each of our labs suggests that this project will be successful. We have a strong team of researchers from three countries with complementary expertise in molecular, morphological, and acoustic analyses. The team includes senior researchers, early- career scientists, and graduate and undergraduate students who will all receive training as part of this project. The longer-term goals of our group are to describe the remaining NZ and Australian cicada species, to construct a phylogenetic tree for the family Cicadidae (3000 species worldwide) that will contribute to the international effort to construct a Tree of Life. Specimens collected as part of the present proposal will include all cicada lineages (for future projects) and other homopteran relatives (for colleagues). Each collecting trip we have made in our preliminary work has turned up 50 to 80 percent undescribed species.
 Undergraduate researcher Rachel Krauss studied the phylogenetics, phylogeography and biogeography of the Australian cicada genus Gudanga whose nine putative species are distributed across arid Australia and are related to the Australian genus Pauropsalta (Marshall, Hill, Vanderpool, and Simon, unpubl). Gudanga species are interesting because they have very subtle (or no) song differences between some of the named species while all other cicada species are distinguished by species-specific calling songs. Gudanga species are mostly described from hind-wing and abdominal color patterns and some of these are recorded as occurring sympatrically. Rachel explored genetic variation within and among species and created a maximum likelihood phylogenetic tree of taxa based on mitochondrial COI and COII gene segments plus nuclear Elongation-Factor-1-alpha sequence. She mapped her tree onto the landscape of Australia and discovered three geographically coherent, cryptic, potential new species. She dissected, cleared, photographed (using Automontage) and drew the genitalia. She described the color patterns and measured each of the putative species identified by the DNA data. Rachel presented her work as a poster at the UCONN campuswide "Frontiers in Undergraduate Research Colloquium" (see photo above).
Undergraduate researcher Rachel Krauss studied the phylogenetics, phylogeography and biogeography of the Australian cicada genus Gudanga whose nine putative species are distributed across arid Australia and are related to the Australian genus Pauropsalta (Marshall, Hill, Vanderpool, and Simon, unpubl). Gudanga species are interesting because they have very subtle (or no) song differences between some of the named species while all other cicada species are distinguished by species-specific calling songs. Gudanga species are mostly described from hind-wing and abdominal color patterns and some of these are recorded as occurring sympatrically. Rachel explored genetic variation within and among species and created a maximum likelihood phylogenetic tree of taxa based on mitochondrial COI and COII gene segments plus nuclear Elongation-Factor-1-alpha sequence. She mapped her tree onto the landscape of Australia and discovered three geographically coherent, cryptic, potential new species. She dissected, cleared, photographed (using Automontage) and drew the genitalia. She described the color patterns and measured each of the putative species identified by the DNA data. Rachel presented her work as a poster at the UCONN campuswide "Frontiers in Undergraduate Research Colloquium" (see photo above).