Phylogenetics: Compositional Heterogeneity Lab

|
EEB 5349: Phylogenetics |
| The goal of this lab is to introduce you to the influence of compositional heterogeneity on phylogeny. |
Compositional heterogeneity means that the equilibrium nucleotide frequencies (or amino acid frequencies for protein data) change across the tree, something that is not accounted for by the standard nucleotide and amino acid models, which assume stationarity (transition probabilities do not change across the tree and one set of equilibrium frequencies applies to every point along any edge of the tree). Non-stationarity can lead to compositional attraction artifacts in which tips with similar nucleotide composition group together even though they may be completely unrelated
Contents
Simulated data and the true tree
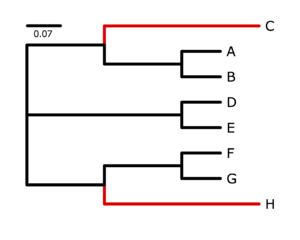
The program p4, written by Peter Foster, specializes in simulating and analyzing data in which nucleotide composition varies across the tree. See Foster (2004) for an introduction to the potential problems created by nucleotide frequency heterogeneity. I used p4 to simulate data on the tree shown on the right. The black-colored lineages were characterized by an AT-biased nucleotide composition very different from the red-colored lineages, which were strongly GC-biased. My goal was to generate a data set that would be very susceptible to nucleotide compositional attraction under ordinary substitution models, and in that I succeeded (as you will see). Taxa C and H share many similarities due to the large number of G and C bases they have independently acquired from their AT-rich ancestors, and it will be very tempting for an ordinary nucleotide model such as GTR to place C and H together.Note: do not use qlogin initially. If you are in a qlogin shell, you will not be allowed to submit a job using qsub (which can only be done from the head node, which is the machine that you are on when you first login to the cluster). So, if you qlogin now, you will need to get out of your qlogin shell using Ctrl-d before running nhPhyloBayes (which uses sneakily uses qsub to submit your job for you).
Download the 500-site simulated data set to a directory named nhlab in your home directory on the cluster:
mkdir nhlab cd nhlab curl http://hydrodictyon.eeb.uconn.edu/people/plewis/courses/phylogenetics/data/sim500nhpb.nex > sim500.nex
At the end of this lab, I will show you how I simulated this data set in p4, but I don't want you to take time doing that now - the analysis will take quite a bit of lab time so I want you to get started on that right away.
Running nhPhyloBayes
You will perform analyses using the program nhPhyloBayes, written by Samuel Blanquart, which implements models described in Blanquart and Lartillot (2006,2008). nhPhyloBayes is a modified version of PhyloBayes (Lartillot, Lepage, and Blanquart, 2009), which is an important Bayesian phylogenetic software package in its own right. nhPhyloBayes is available as a tar archive from nh_PhyloBayes_0.2.3.tar, but I have already downloaded, compiled, and installed nhPhyloBayes for you on the cluster, so you can start your run right away. The following command assumes you are currently inside ~/nhlab, where ~ is a symbol that means your home directory. Note that we are not using qsub to start this run; I'll explain everything in a minute, but for now just (very carefully) type in the following line (or, better yet, copy and paste it) and then press the enter key:
/common/nh/nhpb -d sim500.nex -f sim500 -m bp -x ~/nhlab -y /common/nh/ -Q
Here is a complete explanation of everything you've accomplished on that one line:
- /common/nh/nhpb starts the nhPhyloBayes program. It is located in the /common/nh directory, which is probably not in the path that the system searches to find programs, so you must be explicit and provide the full path
- -d sim500.nex specifies the name of the data file, which should be located in the directory you are in when you issue this command
- -f sim500 specifies the prefix that will be used for all output files (the fact that it is the same as the data file name prefix just illustrates my lack of imagination)
- -m bp specifies the model. nhPhyloBayes does not give you a lot of choices with respect to the model, and bp is the simplest nh (non-homogeneous) model offered by the program
- -x ~/nhlab should be the name of the directory you are in when you issue the command (note: no trailing slash)
- -y /common/nh/ should be the name of the directory where the nhPhyloBayes program is located (note: this time you do add a trailing slash)
- -Q tells nhPhyloBayes to start your program using qsub. So, even though it looked like we avoided using qsub, in reality we used qsub to start the analysis
You should be able to check whether the program is running using the qstat command. If the program did not start, let one of us know and we'll help you get it going. Once you get the program going, we will let it run for about an hour before checking the results.
Ordinary models yield the compositional attraction tree
Before seeing how nhPhyloBayes does with this example, I want you to convince yourself that ordinary models are severely mislead by the strong convergence in nucleotide composition exhibited by these data. Create a nexus file named paup.nex inside your ~/nhlab directory with the following contents:
#nexus begin paup; log file=pauplog.txt start replace; set crit=like; exe sim500.nex; lset nst=6 rmatrix=estimate basefreq=estimate rates=gamma shape=estimate pinvar=estimate; nj; lscores 1; lset rmatrix=previous basefreq=previous shape=previous pinvar=previous; alltrees; savetrees file=gtrig-alltrees.tre brlens; log stop; end;
Answer these questions about this file before you try to run it in PAUP*:
- What will be the name of the file in which PAUP's output will be stored? answer
- What is the optimality criterion that will be used for the search? answer
- Why is a neighbor joining tree generated? answer
- What model is used? answer
- What is the purpose of the second lset command? answer
- What type of search will be conducted? answer
- How many tree topologies will PAUP examine? answer
- In what file will the globally optimum tree be saved? answer
Now run the paup.nex file in PAUP*. Ordinarily you would create a script and feed that script to the qsub command to start the analysis, but it is possible to avoid creating a script by using the concept of a unix pipe - use echo to just write out the command you want to run and pipe this to qsub using the pipe symbol "|" (If you happen to have issued qlogin previously, you will first need to get out of the qlogin shell using Ctrl-d).
echo "/export/apps/paup paup.nex" | qsub -cwd -N naive
Here I've had to be explicit about the location of the paup command (you can find out such full paths to program files using the command which paup) and I've used -N naive to arbitrarily name my job "naive" (to make it easy to find when using qstat to check on what is still running).
PAUP* should only require about a minute or two to complete the exhaustive search. When the "naive" run disappears from qstat output, look for the files pauplog.txt (which contains PAUP's output) and gtrig-alltrees.tre (which contains the best tree). Bring the tree file back to your laptop and view it in FigTree.
- How is this tree different from the true tree? answer
Non-homogeneous models yield the true tree
It is time now to check in on how your nhPhyloBayes run is doing. Use the monitor program (one of about a dozen programs distributed as part of nhPhyloBayes) to see how many samples have accrued since you started it running:
/common/nh/monitor -f sim500
If you are not currently in the directory ~/nhlab, or if you specify the wrong file name prefix after the -f, you will get a strange error message like this:
error while reading MCParameters : do not recognise the program version read -1.-1 instead of 2.3
Assuming you issued the monitor command correctly and from the right place, you should see something like this:
data file sim500a.nex Nsite 500 Ntaxa 8 save every 10 sample size 157 total time : 1 hour, 12 min, 11 sec. time per sample point : 27.585 seconds StatInfCount 0
This tells you that the program has sampled 157 trees in a little over an hour and 12 minutes. The "time per sample point" allows you to estimate how much time it will take to generate larger samples, but due to time constraints we will have to go ahead and generate a consensus tree from the samples we already have. Hopefully you have at least 120 samples by now; if not, wait until you have at least that many before continuing. It doesn't hurt to run the monitor command repeatedly, so predict how much longer you need to wait and check back again at that time.
To use the last 100 trees sampled to create a consensus tree, use the readtopo program as follows (here I'm assuming you just checked and found that you had 157 sampled trees):
/common/nh/readtopo -c -x 57 1 sim500
The -c tells readtopo that you want to create a consensus tree, and the -x 57 says to omit the first 57 trees (leaving 100 for building the consensus tree). The 1 means that you want to use every tree after the first 57 (i.e. if you said 2 it would only use every other tree), and sim500 is the file name prefix you have been using. This should generate four new files, one of which is named sim500_sample.con.tre. Bring this file back to your laptop and open it in FigTree (specify a name for the marginal clade posteriors).
- Does the consensus tree represent the true tree or the nucleotide convergence tree? answer
- What is the lowest marginal posterior probability estimated for any split? answer
FYI: Things I had to do to get nhPhyloBayes to read my NEXUS data file
nhPhyloBayes is extremely picky about some aspects of the nexus file format. Here are three things I had to do in order to get nhPhyloBayes to read the sim500.nex data file:
- Make all nucleotides uppercase letters (TextWrangler can help with this)
- Should be "Matrix", not "matrix"
- Should be "Begin data", not "begin data"
You will probably find more issues when you try to modify your own nexus files. One symptom that you haven't done enough to your file to make nhPhyloBayes happy is that nhPhyloBayes will say something like this (in the case of "matrix" vs "Matrix"):
expected to read "Matrix" terminate called without an active exception Aborted
or this (in the case of a lowercase nucleotide):
taxon 0 site 0 c PhyloParameters::RegisterWithData : error in zip data making error in data matrix : -2
FYI: How I generated sim500.nex
To (re)create the file sim500.nex used in this exercise, create the following python script (p4 is written as an extension to Python):
from p4 import *
func.reseedCRandomizer(13579)
read("( (C:0.4,(A:0.1,B:0.1):0.2):0.2, (D:0.1,E:0.1):0.4, ((F:0.1,G:0.1):0.2,H:0.4):0.2 );")
t = var.trees[0]
a = func.newEmptyAlignment(dataType='dna', taxNames=['A','B','C','D','E','F','G','H'], length=500)
t.data = Data([a])
x = t.newComp(free=1, spec='specified', symbol='A', val=[0.4, 0.1, 0.1, 0.4])
y = t.newComp(free=1, spec='specified', symbol='B', val=[0.01, 0.49, 0.49, 0.01])
t.setModelThing(x, node=t.root, clade=1)
t.setModelThing(y, node=2, clade=1)
t.setModelThing(y, node=13, clade=1)
t.newRMatrix(free=1, spec='2p', val=4.0)
t.setNGammaCat(nGammaCat=4)
t.newGdasrv(free=1, val=0.5)
t.setPInvar(free=1, val=0.0)
t.draw(model=1)
t.simulate(calculatePatterns=True, resetSequences=True, resetNexusSetsConstantMask=True)
t.data.writeNexus(fName="sim500.nex", writeDataBlock=1, interleave=0, flat=1, append=0)
t.data.compoSummary()
The p4 manual describes most of the commands in this file, so I will only go into detail on a few key commands.
- The read command provides a means for creating the true tree used as the basis for simulating data
- The line defining a creates an empty data matrix that we will later fill with simulated data
- The lines defining x and y create new compositions (sets of nucleotide frequencies) which we will assign to different sections of the tree before simulating data
- The setModelThing lines assign composition x to most of the tree and composition y to just 2 edges (you can use t.draw() to find out what numbers are assigned to each node)
- The simulate command actually simulates the data
- The writeNexus command stores the simulated data in the file sim500.nex
- The last line shows a summary of the nucleotide composition of each taxon
Simulating just one data set takes so little time that you can just run this file directly - no need to use qsub:
/opt/python/bin/python dosim.py
Here, dosim.py is the name of the Python script you just created, and note that I used the full path to python. The reason that the full path is needed is that there is more than one python version installed on the cluster (this one is 2.7.3, while the default one is 2.6.6), and only version 2.7.3 has p4 installed. In fact, if you want to always use this version (a good idea), add this line to the bottom of your ~/.bashrc file:
export PATH="/opt/python/bin:$PATH"
The ~/.bashrc file is read each time you login, and the line we added causes /opt/python/bin to be inserted at the beginning of your PATH, which is an environmental variable that tells the operating system where to look for programs that you ask it to run. Now when you ask for python, it will find the python in /opt/python/bin before it finds the default version, which is in /usr/bin. You can add the line above either by editing ~/.bashrc in nano, or you can append the line using the cat command (be sure that the hypthen and ">>" each are separated by spaces, and there is no space between the two ">" symbols):
cat - >> ~/.bashrc export PATH="/opt/python/bin:$PATH" Ctrl-d
After entering the first line above, the cat command will sit and wait for you to type the second line. After entering the second line, press (don't type out) Ctrl-d - this is a signal to the cat command that you have finished entering the text that you want to append to the file ~/.bashrc. The symbol >> means "append" whereas just ">" would mean "overwrite", so be careful to use ">>" if you go this route.
Literature Cited
Blanquart S., Lartillot N. 2006. A Bayesian compound stochastic process for modeling nonstationary and nonhomogeneous sequence evolution. Molecular Biology and Evolution. 23:2058–2071.
Blanquart S., Lartillot N. 2008. A Site- and Time-Heterogeneous Model of Amino Acid Replacement. Molecular Biology and Evolution. 25:842–858.
Foster P.G. 2004. Modeling Compositional Heterogeneity. Systematic Biology. 53:485–495.
Lartillot N., Lepage T., Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 25:2286–2288.