Phylogenetics: r8s Lab

|
EEB 349: Phylogenetics |
| The goals of this final lab are to learn how to: (1) set up a NEXUS file for the r8s program; (2) interpret the most important parts of the output of r8s; and (3) view a tree with divergence times produced by r8s in TreeView and FigTree. |
Contents
- 1 Using r8s to estimate divergence times
- 2 Start the program PuTTY and connect to bbcxsrv1.biotech.uconn.edu
- 3 Inspecting SAMPLE_SIMPLE
- 4 Adding a node of interest using the MRCA command
- 5 Running r8s on SAMPLE_SIMPLE
- 6 Viewing the results
- 7 Using cross-validation to determine optimum smoothing
- 8 PL analysis using the optimum penalty weighting
- 9 Using TreeView to visualize/print/copy trees
- 10 Using FigTree to visualize/print/copy trees
Using r8s to estimate divergence times
There is no version of r8s for the Windows operating system, so we will use the version installed on the Bioinformatics cluster instead (the machines composing the cluster are all MacIntoshes running OS X).
Start the program PuTTY and connect to bbcxsrv1.biotech.uconn.edu
If you do not find PuTTY on your desktop, you can download the program again from the PuTTY website. Using PuTTY, connect to bbcxsrv1.biotech.uconn.edu.
Inspecting SAMPLE_SIMPLE
r8s is installed on the cluster, so we don't need the program itself, but we do need the sample data file that comes with r8s for the lab today. I've placed the SAMPLE_SIMPLE file that comes with r8s on a web server, and you can download it directly to the cluster using the curl program. Note that you could download the program using a web browser, then transfer the file to the cluster using PSFTP (or some other SFTP program), but using curl is easier and faster:
First, create a directory for this lab in your home directory:
mkdir $HOME/r8slab cd $HOME/r8slab
Now, download the SAMPLE_SIMPLE data file there:
curl -o SAMPLE_SIMPLE http://hydrodictyon.eeb.uconn.edu/eeb349/SAMPLE_SIMPLE
Use the pico editor to see the first few lines of the SAMPLE_SIMPLE file:
pico SAMPLE_SIMPLE
Note that r8s uses the NEXUS format for its input files. Remember that to exit pico, you need to press the key combination Ctrl-x.
See if you can answer these questions just by looking at the SAMPLE_SIMPLE file:
- Which NEXUS blocks are present? (Hint: you should find 2 NEXUS blocks in this file)
- Why does r8s need to know the number of sites? (as in the "blformat nsites =952 lengths=persite" command)
Adding a node of interest using the MRCA command
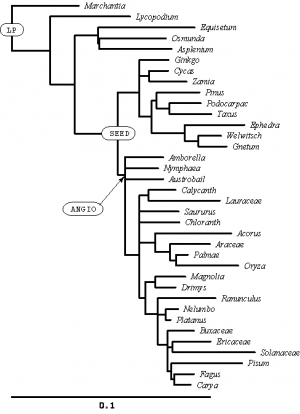
In the SAMPLE_SIMPLE file, these two lines define names for two interior nodes of the tree:
mrca LP marchantia pisum; mrca ANGIO amborella pisum;
I have labeled these nodes on the tree (right). You might need to click on the figure to expand it before you will be able to read the text. Can you see how these simple mrca (Most Recent Common Ancestor) statements uniquely define interior nodes?
Create an mrca statement to create a name for the node I have labeled SEED (there are many different ways you could do this, all of which would uniquely specify the labeled node). Add your statement in the SAMPLE_SIMPLE file beneath the other two using the pico editor.
- Which line in the data file serves to calibrate the (relaxed) clock?
Running r8s on SAMPLE_SIMPLE
Create a file (using pico) named gor8s that will serve as your qsub script for running r8s:
#$ -o simple.txt -j y cd $HOME/r8slab r8s -b -f SAMPLE_SIMPLE
The #$ -o simple.txt -j y on the first line is an instruction that programs composing the Sun Grid Engine software pay attention to. Normally, in unix scripts, if a pound (hash) symbol (i.e. #) is the first thing on a line, the entire line is treated as a comment and ignored. Programs such as qsub (part of the Sun Grid Engine) also ignore lines beginning with just a hash symbol, but they do pay attention to lines that directly follow the hash symbol with a dollar sign. In this case, the #$ -o simple.txt -j y on the first line is interpreted as a command by qsub. The command instructs qsub to save the output (-o) of whatever program is being run to a file named simple.txt. The -j y tells it to join (-j) any error output to the same file (simple.txt) rather than creating a separate file just to report errors. The y just means "yes", as in "yes, do join error to the other output".
The second line, cd $HOME/r8slab, says to change directories (cd) into the r8slab subdirectory of your home directory. $HOME is an "environmental" variable that is always guaranteed to expand to the full path to your home directory. You can type the command env if you want to see the values of all environmental variables currently defined. The value of any of these can be used in script by prepending the variable name with the dollar sign. Note that if your gor8s script is in your home directory, you could just say cd r8slab rather than cd $HOME/r8slab. Being explicit, however, about the location of the r8slab directory means that you could move your gor8s script anywhere and it would still work the same way.
The -f option tells r8s that the data file name is coming next, and the -b option causes r8s to run in batch mode (which simply means it will execute the specified data file and then quit, rather than producing prompts and waiting for answers).
Ok, run your r8s analysis using the qsub command:
qsub gor8s
This run will take almost no time at all. In fact, if you type qstat immediately after your qsub command, you may not see your run because it will have already finished!
You should look for the output (simple.txt) in your home directory. It is possible to tell qsub to save simple.txt in your r8slab subdirectory by making the first line of your gor8s script look like this:
#$ -o r8slab/simple.txt -j y
Note that you cannot, however, use $HOME (or other environmental variables) in this line because qsub doesn't know how to deal with that. Just remember that the default behavior of qsub is to save your output file to your home directory.
Viewing the results
Open simple.txt in pico and take a look at the output that r8s generated. You can ignore the following warning, which appears on the first couple of lines
Warning: no access to tty (Bad file descriptor). Thus no job control in this shell.
The reason for this warning is explained here, but it is easier to simply ignore the warning than to prevent it!
There is a lot of output here, so skip down to the interesting part, which is a section labeled Estimated ages and substitution rates for tree PAUP_9. This shows a big table with a line showing the age for every node in the tree.
- What method did r8s use here? (in lecture I mentioned four methods that were implemented in this program; in this case it is using the least sophisticated of these four)
- What is the age of the node labeled LP? Is this age surprising, or could you have predicted this before running r8s?
- What is the age of the seed plants? (look for the node you named using your mrca command)
Using cross-validation to determine optimum smoothing
Let's run one more example to illustrate the Penalized Likelihood (PL) approach with cross-validation. Download the file SAMPLE_CROSSVAL using curl:
curl -o SAMPLE_CROSSVAL http://hydrodictyon.eeb.uconn.edu/eeb349/SAMPLE_CROSSVAL
then modify your gor8s file to look like this (note that both the first and third lines were changed):
#$ -o r8slab/crossval.txt - j y cd $HOME/r8slab r8s -b -f SAMPLE_CROSSVAL
Run your gor8s file using qsub. This should generate the file crossval.txt inside your r8slab subdirectory. Before looking at the output, look at the SAMPLE_CROSSVAL file itself. The part of this file that is most different from SAMPLE_SIMPLE is near the end:
divtime method=pl algorithm=tn cvStart=0 cvInc=0.5 cvNum=8 crossv=yes;
The divtime command will do cross-validation for 8 values of the weighting parameter (which I referred to as lambda in lecture). This parameter determines how much weight to give to the penalty function compared to the likelihood function.
- Which 8 values of the weighting parameter will be tried?
Now look at the output saved in the file crossval.txt
- Can you determine which value of the weighting parameter (r8s refers to this as the smoothing parameter) is best? Look for the smallest value in the last ("Chi Square Error") column, then choose the value from the first ("log10 smooth") column in that same row.
PL analysis using the optimum penalty weighting
Now put together a new analysis using the optimum penalty weight derived from the cross validation analysis. Start by making a copy of the SAMPLE_CROSSVAL file, naming the copy myfile.nex:
cp SAMPLE_CROSSVAL myfile.nex pico myfile.nex
Once you have myfile.nex open in pico, comment out the last two commands (set and divtime) in the r8s block, and add your own divtime command to do the analysis one more time using the optimal value for the penalty weight:
#NEXUS . . . collapse; fixage taxon=lp age=420; set ftol=1e-7 maxiter=2000; set verbose=1; [suppresses huge amount of output in CV analyses] [divtime method=pl algorithm=tn cvStart=0 cvInc=0.5 cvNum=8 crossv=yes;] set smoothing=2.5; divtime method=pl algorithm=tn; describe plot=tree_description; end;
Note that I've added a describe command (after divtime and before end) to get a tree description with branch lengths expressed as times.
Remember to modify your gor8s script to account for the fact that you want to run myfile.nex this time rather than SAMPLE_CROSSVAL:
#$ -o r8slab/myfile.txt - j y cd $HOME/r8slab r8s -b -f myfile.nex
Use qsub to start the analysis.
Using TreeView to visualize/print/copy trees
The next step will be to copy the tree description to a file on your local computer and open the file in Rod Page's TreeView program to view it. TreeView is a free program available for MacIntosh, Windows and Linux operating systems. The first step is to create an empty file into which we will paste the tree description. To do this, right-click on some empty space in your directory (on your local PC machine). From the menu that pops up, choose New > Text Document. Rename this newly created file r8s.tre. Open the file for editing in PAUP* (or any another program that can edit plain text files: for example, the Notepad program that comes with Windows will work).
Back on the cluster, in the output of r8s after you run the above file, there should be a tree description at the very end. Use the cat command to show this tree description to you in the PuTTY console window (cat just displays the entire file):
cat myfile.txt
Select the tree description (start selecting at the first left parenthesis and include the semicolon at the end, but leave out the "tree PAUP_1 = " part). Once it is selected, press either Ctrl-C or Ctrl-Ins to copy it to the clipboard.
Now paste the tree description into your r8s.tre document and save the file.
Because we provided a name for this file that ends in ".tre", TreeView will recognize it as a tree file and TreeView will start automatically when the file is double-clicked. Do that now.
When TreeView opens, you should see the tree in cladogram view. To see divergence times appropriately scaled, choose Tree > Phylogram from TreeView's main menu. Note that the tree is ultrametric (the length of a path from an ancestor to any descendant is the same value). The scale bar at the bottom left is calibrated in millions of years.
Choosing Tree > Show internal edge labels will reveal internal nodes that were given labels. Each of these corresponds to one mrca command.
Note that you can use Ctrl-C to copy a tree from TreeView in a form that can be pasted into a graphics program such as Microsoft Powerpoint, Adobe Illustrator, or Microsoft Word. You can also print directly from TreeView, but the results are less than spectacular. I usually paste it into Illustrator and clean it up a bit before printing or saving it as a PDF file.
Using FigTree to visualize/print/copy trees
A recent entry into the field of phylogenetic tree viewers is a program called FigTree, by Andrew Rambaut (who has also authored or co-authored a host of other very nice software applications for phylogenetics, including Beast, Tracer and Seq-Gen). FigTree is not installed on your PC, so you will have to do that first.
Downloading and installing FigTree
- Click the Download button labeled "Windows executable version" on the right side of the FigTree web page.
- Save the zip file to your desktop. Note that the zip file looks like a folder, but the zipper in the icon tells you that it is actually a compress zip archive. Don't try to dive into this apparent folder; extract the files therein first (next step).
- Extract the files by right-clicking on the name of the zip file (FigTree v1.0.zip) on your desktop. This should create a FigTree v1.0 folder.
Running FigTree
I will assume you still have your r8s.tre file from the previous (TreeView) section. If not, go back to that section and re-create it.
Start FigTree by going into the FigTree v1.0 folder twice (there is another FigTree v1.0 folder inside the one on your desktop). Inside the inner folder, you should find the FigTree v1.0.exe file. Double-click this to start FigTree, then choose your r8s.tre file when asked for a tree file to open.
After a pause (which may be a long one depending on the computer you are using), FigTree will open its main window and show you the tree.
Increasing size of taxon labels
You will notice that the taxon names are very small. You can fix that by expanding the Tip Labels section in the left panel and increasing the font size.
Showing divergence times
Now click the checkbox beside Node Labels, then expand that panel. Increase the font size to 12 and decrease the Sig. digits to 2.
Increasing line thickness
You might also want to increase the thickness of the lines making up the tree. You can do this by expanding the Appearance panel, then increasing Line Width.
Saving tree as a PDF file
Now you can create a PDF file by either clicking Ctrl-E or choosing File > Export PDF... from the main menu of FigTree. Be sure to add the .pdf extension on the file name you choose, otherwise Adobe Acrobat Reader will not recognize it as a PDF file.
Feel free to play with all the other gadgets available to you in FigTree.