Difference between revisions of "Phylogenetics: r8s Lab"
(→Download r8s) |
(→Running r8s on SAMPLE_SIMPLE) |
||
| Line 57: | Line 57: | ||
The <tt>-f</tt> option tells r8s that the data file name is coming next, and the <tt>-b</tt> option causes r8s to run in batch mode (which simply means it will execute the specified data file and then quit, rather than producing prompts and waiting for answers). | The <tt>-f</tt> option tells r8s that the data file name is coming next, and the <tt>-b</tt> option causes r8s to run in batch mode (which simply means it will execute the specified data file and then quit, rather than producing prompts and waiting for answers). | ||
| + | |||
| + | '''BEGIN AGAIN HERE''' | ||
The "#$ -o simple.txt -j y" on the first line | The "#$ -o simple.txt -j y" on the first line | ||
Revision as of 02:03, 20 April 2007

|
This article is still under construction. Expect it to change frequently until this notice is removed. |

|
EEB 349: Phylogenetics |
| The goals of this final lab are to learn how to: (1) set up a NEXUS file for the r8s program; (2) interpret the most important parts of the output of r8s; (4) view a tree with divergence times produced by r8s in TreeView. |
Contents
Using r8s to estimate divergence times
There is no version of r8s for the Windows operating system, so we will use the version installed on the Bioinformatics cluster instead.
Start the program PuTTY and connect to bbcxsrv1.biotech.uconn.edu
If you do not find PuTTY on your desktop, you can download the program again from the PuTTY website. Using PuTTY, connect to bbcxsrv1.biotech.uconn.edu.
Download r8s
r8s is installed on the cluster, so we don't need the program itself, but we do need the sample data file that comes with r8s for the lab today. I've placed the SAMPLE_SIMPLE file that comes with r8s on a web server, and you can download it directly to the cluster using the curl program. Note that you could download the program using a web browser, then transfer the file to the cluster using PSFTP (or some other SFTP program), but using curl is easier and faster:
First, create a directory for this lab in your home directory:
mkdir $HOME/r8slab cd $HOME/r8slab
Now, download the SAMPLE_SIMPLE data file there:
curl -o SAMPLE_SIMPLE http://hydrodictyon.eeb.uconn.edu/eeb349/SAMPLE_SIMPLE
Use the more command to see the first few lines of the SAMPLE_SIMPLE file. Note that r8s uses the NEXUS format for its input files. You can see more of the file by pressing the spacebar, and you can quit viewing the file by pressing the letter q (for quit).
See if you can answer these questions just by looking at the SAMPLE_SIMPLE file:
- Which NEXUS blocks are present? (Hint: you should find 2 NEXUS blocks in this file)
- Why does r8s need to know the number of sites? (as in the "blformat nsites =952 lengths=persite" command)
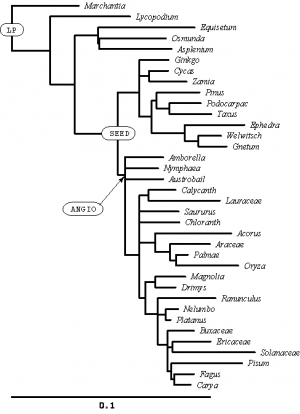
In the SAMPLE_SIMPLE file, these two lines define names for two interior nodes of the tree:
mrca LP marchantia pisum; mrca ANGIO amborella pisum;
I have labeled these nodes on the tree (right). You might need to click on the figure to expand it before you will be able to read the text. Can you see how these simple mrca (Most Recent Common Ancestor) statements can uniquely define interior nodes?
Create an mrca statement to create a name for the node I have labeled SEED (there are many different ways you could do this, all of which would uniquely specify the labeled node).
Add your statement in the SAMPLE_SIMPLE file beneath the other two using the pico editor:
pico SAMPLE_SIMPLE
The command above will invoke the pico text editor program and you should see the contents of the file displayed in the main editor window. Use the arrow keys to navigate down to the two existing mrca lines, then add your mrca command after these. To exit pico, use the Ctrl-X key combination (the ^ symbol in the hints at the bottom of the window refers to the Ctrl key), then answer Y when asked if you want to save the modified buffer, and finally just press the Enter key when asked "File name to write : SAMPLE_SIMPLE"
- Which line in the data file serves to calibrate the (relaxed) clock?
Running r8s on SAMPLE_SIMPLE
Create a file (using pico) that will serve as your qsub script for running r8s:
#$ -o simple.txt -j y cd $HOME/r8slab r8s -b -f SAMPLE_SIMPLE
The -f option tells r8s that the data file name is coming next, and the -b option causes r8s to run in batch mode (which simply means it will execute the specified data file and then quit, rather than producing prompts and waiting for answers).
BEGIN AGAIN HERE
The "#$ -o simple.txt -j y" on the first line
alleyn [/home/tmp1/dist/sample] 13% more simple.txt
There is a lot of output here, so skip down to the interesting part, which is a section labeled "Estimated ages and substitution rates for tree PAUP_9". This shows a big table with a line showing the age for every node in the tree.
What method did r8s use here? (in lecture I mentioned four methods that were implemented in this program)
What is the age of the node labeled LP? Is this age surprising, or could you have predicted this before running r8s?
What is the age of the seed plants? (look for the node you named using your mrca command)
Using cross-validation to determine optimum smoothing
Let's run one more example to illustrate PL with cross-validation:
alleyn [/home/tmp1/dist/sample] 14% r8s -b -f SAMPLE_CROSSVAL > crossval.txt
Before looking at the output, look at the SAMPLE_CROSSVAL file itself (use pico if you want). The part of this file that is most different from SAMPLE_SIMPLE is near the end:
divtime method=pl algorithm=tn cvStart=0 cvInc=0.5 cvNum=8 crossv=yes;
Which of the four methods mentioned in lecture is being used now?
The divtime command will do cross-validation for 8 values of the weighting parameter (determines how much weight to give to the penalty function compared to the likelihood function).
Which 8 values of the weighting parameter will be tried?
Now look at the output saved in the file crossval.txt, using either the more command or the pico editor.
Can you determine which value of the weighting parameter is best? PL analysis using the optimum penalty weighting Now put together a new analysis using the optimum penalty weight derived from the cross validation analysis. Let's simply modify the SAMPLE_CROSSVAL file:
alleyn [/home/tmp1/dist/sample] 15% cp SAMPLE_CROSSVAL myfile.nex alleyn [/home/tmp1/dist/sample] 16% pico myfile.nex
Once you have myfile.nex open in pico, comment out the last two commands (set and divtime) in the r8s block, and add your own divtime command to do the analysis one more time using the optimal value for the penalty weight:
#NEXUS . . . collapse; fixage taxon=lp age=420; set ftol=1e-7 maxiter=2000; [set verbose=0;] [suppresses huge amount of output in CV analyses] [divtime method=pl algorithm=tn cvStart=0 cvInc=0.5 cvNum=8 crossv=yes;] set smoothing=2.5; divtime method=pl algorithm=tn; end;
Also add a command (after divtime and before end) to get a tree description with branch lengths expressed as times (look at SAMPLE_SIMPLE for an example of how to do this). Using TreeView to visualize/print/copy trees We will now copy the tree description to a file on your local computer and open the file in Rod Page's TreeView program to view it. TreeView is a free program available (from here) for MacIntosh, Windows and Linux operating systems. The first step is to create an empty file into which we will paste the tree description. To do this, right-click on some empty space in your directory. From the menu that pops up, choose New > Text Document. Rename this newly created file r8s.tre. Double-click the file to open it.
In the output of r8s after you run the above file, there should be a tree description at the very end. Use the cat command to show this tree description to you in the PuTTY console window (cat just displays the entire file, not stopping like more).
Select the tree description (start selecting at the first left parenthesis and include the semicolon at the end, but leave out the "tree PAUP_1 = " part). Once it is selected, press either Ctrl-C or Ctrl-Ins to copy it to the clipboard.
Now click the title bar of the window containing your new, empty text file. Once that window has the input focus, Ctrl-V to paste in the tree description. Close the window and save the file.
Because we provided a name for this file that ends in ".tre", TreeView will recognize it as a tree file and TreeView will start automatically when the file is double-clicked. Do that now.
When TreeView opens, you should see the tree in cladogram view. To see divergence times appropriately scaled, choose Tree > Phylogram from TreeView's main menu.
Note that you can use Ctrl-C to copy a tree from TreeView in a form that can be pasted into a graphics program such as Powerpoint, Illustrator, or Word. You can also print directly from TreeView, but the results are less than spectacular. I usually paste it into Illustrator and clean it up a bit before printing or saving it as a PDF file.