Difference between revisions of "Phylogenetics: RevBayes Lab"
Paul Lewis (Talk | contribs) (→Relaxed clocks) |
Paul Lewis (Talk | contribs) (→Reviewing results of the relaxed clock analysis) |
||
| Line 286: | Line 286: | ||
rb relaxed.Rev | rb relaxed.Rev | ||
| − | == | + | == Review results of the relaxed clock analysis == |
| + | |||
| + | If you create a plot of your ''relaxed.dot'' file using one of the online Graphviz viewers, the increase in model complexity will be made very apparent! | ||
| + | |||
| + | Open the ''relaxed.log'' file in Tracer and think about the following questions before peeking at the answer: | ||
Revision as of 22:27, 20 April 2020

|
EEB 5349: Phylogenetics |
| The goal of this lab exercise is to introduce you to Bayesian divergence time estimation using RevBayes. There are other programs that are currently more popular than RevBayes for doing this (notably BEAST2), but I prefer RevBayes for this lab because it is less of a black box: every aspect of the model is explicitly defined in RevBayes. |
Contents
Getting started
Login to Xanadu
Login to Xanadu and request a machine as usual:
srun --pty -p mcbstudent --qos=mcbstudent bash
Once you are transferred to a free node, load the paml, paup, and revbayes modules
module load paml/4.9 module load paup/4.0a-166 module load RevBayes/xxx
Create a directory
Use the unix mkdir command to create a directory to play in today:
cd ~ # you can omit this line if you are already in your home directory mkdir rblab
Simulating and analyzing under the strict clock model
Divergence time analyses are the trickiest type of analysis we will do in this course. That's because the sequences do not contain information about substitution rates or divergence times per se; they contain information about the number of substitutions that have occurred, and the number of substitutions is the product of rate and time. Thus, maximum likelihood methods cannot separate rates from times; doing so requires a Bayesian approach and considered use of priors, which constrain the range of rate and time scenarios considered plausible.
We will thus start slowly, and we will simulate data so that we know the truth. This will help guide your expectations when conducting divergence time analyses on real data.
PAML evolver
Let's use the evolver program, which is part of Ziheng Yang's PAML package, to simulate data for 10000 sites on a 20-taxon pure birth (Yule) tree using a strict clock. This will allow us to know everything: the birth rate of the tree generating process, the "clock" rate (i.e. the substitution rate that applies to the entire tree), as well as the model used for simulation.
We will each use a different random number seed, so we should all get slightly different answers.
Simulate a tree
First simulate a pure birth tree using evolver. Start evolver by simply typing evolver at the bash prompt, then enter the information provided below at the prompts (for questions that ask for multiple quantities, just separate the values by a space):
- specify that you want to generate a rooted tree by typing 2
- specify 20 species
- specify 1 tree and a random number seed of your choosing
- specify 1 to answer yes to the question about wanting branch lengths
- specify 2.6 for the birth rate, 0.0 for the death rate, 1.0 for the sampling fraction, and 1.0 for the tree height
- press 0 to quit
One thing to note before we continue. PAML's evolver program scales the tree to have height equal to the specified mutation rate (1.0, the last number we specified above). Normally pure birth trees would have different heights because of stochastic variation, but apparently this is only possible in evolver by editing the source code and making your own, ad hoc version. I've done the next best thing, which is set the birth rate to the value (2.6) that yield a tree of expected height 1.
You should now find a tree description in the file evolver.out. Rename this file tree.txt (this will prevent your tree description from being overwritten when you run evolver again, and we will use the tree.txt file as input to RevBayes):
mv evolver.out tree.txt
Simulate sequences
The PAML evolver program requires a control file specifying everything it needs to know to perform your simulation. Create a file named control.dat with the following contents (2 lines require modification: seed and tree description):
2 seed goes here 20 10000 1 -1 tree description goes here 4 5 0 0 0.1 0.2 0.3 0.4
Here's what each of those lines does (consult the evolver section of the PAML manual for more info about each option):
- line 1: 2 specifies that we want the output as a nexus file
- line 2: you should enter your own random number seed on the second line (can be the same as the one you used for the tree)
- line 3: 20 taxa, 10000 sites, 1 data set
- line 4: -1 says to use the branch lengths in the tree description
- line 5: tree description: paste in the tree description you generated from the first evolve run here
- line 6: 4 specifies the HKY model
- line 7: set kappa equal to 5
- line 8: set the gamma shape parameter to 0 and the number of rate categories to 0 (i.e. no rate heterogeneity)
- line 9: set state frequencies to: T=0.1, C=0.2, A=0.3, and G=0.4 (note, not in alphabetical order!)
When saving simulated data in nexus format, PAML's evolver command looks for three files (paupstart, paupend, and paupblock) specifying what text should go at the beginning, end, and following each data matrix generated, respectively. The only one of these files that needs to have anything in it is paupstart. Here's the quick way to create these files:
echo "#nexus" > paupstart touch paupblock touch paupend
The echo command parrots what you put in quotes and the > paupstart at the end creates the file paupstart and saves the echoed contents there (you could use >> paupstart if you wanted to append other lines to the file). The touch command is intended to update the time stamp on a file, but will create an empty text file if the file specified does not exist.
Run evolver now using this control file, and selecting option (5) from the menu, which is "Simulate nucleotide data sets".
evolver 5 control.dat
If you get Error: err tree... it means that you did not follow the directions above ;)
You should now find a file named mc.nex containing the sequence data.
Use RevBayes to estimate the birth rate and clock rate
In our first RevBayes analysis, we will see how well we can estimate what we already know to be true about the evolution of both the tree and the sequences. You will cheat and fix some things to their known true values, such as the tree topology and edge lengths. The idea is to take small steps so that we know what we are doing all along.
RevBayes uses an R-like language called the Rev Language to specify the model and the analysis. Rev is not R, but it is so similar to R that you will often forget that you are not using R and will try things that work in R but do not work in Rev - just a heads-up!
Set up the tree submodel
Create a new file named strict.Rev and add the following to it: I'll provide some explanation below the code block.
# Load data and tree
D <- readDiscreteCharacterData(file="mc.nex")
n_sites <- D.nchar()
T <- readTrees("tree.txt")[1]
n_taxa <- T.ntips()
taxa <- T.taxa()
# Initialize move (nmoves) and monitor (nmonitors) counters
nmoves = 1
nmonitors = 1
# Birth-death tree model
death_rate <- 0.0
birth_rate ~ dnExponential(0.01)
birth_rate.setValue(1.0)
diversification := birth_rate - death_rate
moves[nmoves++] = mvSlide(birth_rate, delta=1.0, tune=true, tuneTarget=0.4, weight=1.0)
sampling_fraction <- 1.0
root_time <- T.rootAge()
timetree ~ dnBDP(lambda = birth_rate,
mu = death_rate,
rho = sampling_fraction,
rootAge = root_time,
samplingStrategy = "uniform",
condition = "nTaxa",
taxa = taxa)
timetree.setValue(T)
Note that we are assigning only the first tree in trees.txt to the variable T (there is only 1 tree in that file, but RevBayes stores the trees it reads in a vector, so you have to add the [1] to select the first anyway).
The functions beginning with dn (e.g. dnExponential and dnBDP) are probability distributions. Thus, birth_rate is a parameter that is assigned an Exponential prior distribution having rate 0.01, and timetree is a parameter representing the tree and its branching times that is assigned a Birth Death Process (BDP) prior distribution. The BDP is a submodel, like the +I or +G rate heterogeneity submodels: it has its own parameters (lambda, mu, rho, and rootAge) all of which are fixed except for birth_rate.
The setValue function sets the starting value of a parameter that is allowed to vary.
Each parameter in the model requires a mechanism to propose changes to its value. These are called moves. A vector of moves has been created for you, so you need only add to it. The variable nmoves keeps track of how many moves we've defined. Each time a move is added to the moves vector, we increment the variable nmoves so that new moves will not overwrite previously defined moves. This increment is performed by the ++ in nmoves++. The fact that the ++ follows nmoves means that nmoves will be incremented after its value is used. If we had used ++nmoves instead, nmoves would have been incremented and then used, which would be incorrect because vector indices in Rev Language, like R, start at 1, not 0.
Monitors in RevBayes handle output. We are just initializing the monitor counters now; we will add monitors toward the end of our RevBayes script.
The model comprises a DAG (Directed Acyclic Graph). The nodes of this graph are of several types and represent model inputs and outputs:
- Stochastic nodes are exemplified by birth_rate and timetree; they can be identified by the tilde (~) symbol used to assign a prior distribution.
- Constant nodes are exemplified by death_rate, sampling_fraction, and root_time; they can be identified by the assignment operator <- that fixes their value to a constant.
- Deterministic nodes are exemplified by diversification; they can be identified by the assignment operator :=. These nodes represent functions of other nodes used to output quantities in a more understandable way. For example, diversification will show up as a column in the output even though it is not a parameter of the model itself. (The diversification node was only added here to illustrate deterministic nodes; it's value will always equal birth_rate because death_rate is a constant 0.0).
I will show you how to create the DAG graphically (ha!) in the form of a pdf before we run the model.
Set up the strict clock submodel
Add the following 3 lines to your growing revscript:
# Strict clock clock_rate ~ dnExponential(0.01) clock_rate.setValue(1.0) moves[nmoves++] = mvSlide(clock_rate, delta=1.0, tune=true, tuneTarget=0.4, weight=1.0)
This adds a parameter clock_rate with a vague Exponential prior (rate 0.01) and starting value 1.0. The move we're using to propose new values for this parameter as well as the birth_rate parameter is a sliding window move, which you are familiar with from your MCMC homework. The value delta is the width of the window centered over the current value, and we've told RevBayes to tune this proposal during the burnin period so that it achieves (if possible) an acceptance rate of 40%. The weight determines the probability that this move will be tried. At the start of the MCMC analysis, RevBayes sums the weights of all moves you've defined and uses the weight divided by the sum of all weights as the probability of selecting that particular move next.
Set up the substitution submodel
Now let's set up a GTR substitution model:
# GTR model state_freqs ~ dnDirichlet(v(1,1,1,1)) exchangeabilities ~ dnDirichlet(v(1,1,1,1,1,1)) Q := fnGTR(exchangeabilities, state_freqs) moves[nmoves++] = mvDirichletSimplex(exchangeabilities, alpha=10.0, tune=true, weight=1.0) moves[nmoves++] = mvDirichletSimplex(state_freqs, alpha=10.0, tune=true, weight=1.0)
The Q matrix for the GTR model involves state frequencies and exchangeabilities. I've made bothstate_freqs and exchangeabilities stochastic nodes in our DAG and assigned both of them flat Dirichlet prior distributions (the v(1,1,...,1) part is the vector of parameters for the Dirichlet prior distribution (all 1s means a flat prior).
I've assigned mvDirichletSimplex moves to both of these parameters. A simplex is a set of coordinates that are constrained to sum to 1, and this proposal mechanism modifies all of the state frequencies (or exchangeabilities) simultaneously while preserving this constraint. A list of all available moves can be found in the Documentation section of the RevBayes web site if you want to know more.
Finalize the PhyloCTMC
It is time to collect the various submodels (timetree, Q, and clock_rate) into one big Phylogenetic Continuous Time Markov Chain (dnPhyloCTMC) distribution object and attach (clamp) the data matrix D to it.
# PhyloCTMC
phySeq ~ dnPhyloCTMC(tree=timetree, Q=Q, branchRates=clock_rate, nSites=n_sites, type="DNA")
phySeq.clamp(D)
mymodel = model(exchangeabilities)
mymodel.graph("strict.dot", TRUE, "white")
The next-to-last line is a little obscure. RevBayes needs to have an entry point (a root node, if you will) into the DAG, and any stochastic node will suffice. Here I've supplied exchangeabilities when constructing mymodel, but I could have provided <state_freqs>, <birth_rate>, <clock_rate>, etc., instead.
The last line creates a file named strict.dot that contains code (in the dot language) for creating a plot of your DAG. The second argument (TRUE) tells the graph command to be verbose, and the last argument ("white") specifies the background color for the plot.
Set up monitors
Let's create 2 monitors to keep track of sampled parameter values, sampled trees, and screen output:
# Monitors monitors[nmonitors++] = mnModel(filename = "output/strict.log", printgen = 10, separator = TAB) monitors[nmonitors++] = mnFile(filename = "output/strict.trees", printgen = 10, timetree) monitors[nmonitors++] = mnScreen(printgen=100)
The first monitor will save model parameter values to a file named strict.log in the output directory (which will be created if necessary). The second monitor will save trees to a file named strict.trees in the output directory. Note that we have to give it timetree as an argument. This is kind of silly because we've fixed the tree topology and edge lengths, so all the lines in the output.strict.trees will be identical, but this saves me having to explain this later. Finally, the third monitor produces output to the console so that you can monitor progress.
Note that we are sampling only every 10th iteration for the first 2 monitors and every 100th iteration for the screen monitor.
Set up MCMC
Finally, we're ready to add the final section to our revscript. Here we create an mcmc object that combines the model, monitors, and moves and says to do just 1 MCMC analysis. We will devote the first 1000 iterations to burnin, stoping to tune the moves every 100 iterations (RevBayes collects data for 100 iterations to compute the acceptance probabilities for each move, then uses that to decide whether to make the move bolder or more conservative.) Then we run for real for 10000 iterations and ask RevBayes to output an operator summary, which will tell us how often each of our moves was attempted and succeeded.
# MCMC mymcmc = mcmc(mymodel, monitors, moves, nruns=1) mymcmc.burnin(generations=1000, tuningInterval=100) mymcmc.run(generations=10000) mymcmc.operatorSummary() quit()
Run RevBayes
To run RevBayes, just type rb at the command prompt followed by the name of your revscript file:
rb strict.Rev
If this were a long analysis, we would create a slurm script and submit the job using sbatch, but this one should be short enough that you can easily wait for it to finish while logged in.
Reviewing the strict clock results
First, copy the contents of the file strict.dot and paste them into one of the online Graphviz viewers such as GraphvizFiddle, GraphvizOnline, or WebGraphviz. The resulting plot shows your entire model as a graph, with constant nodes in square boxes, stochastic nodes in solid-line ovals, and deterministic nodes in dotted-line ovals.
Now download the file strict.log stored in the output directory to your laptop and open it in Tracer.
- What is the 95% HPD (highest posterior density) credible interval for clock_rate (use the Estimates tab in Tracer to find this information)?' Does this include the true value?' answer
- What is the 95% HPD (highest posterior density) credible interval for birth_rate?' Does this include the true value?' answer
- Using the Marginal Density tab in Tracer, and selecting all 6 exchangeabilities, do these densities make sense given what you know about the model used? answer
- Using the Marginal Density tab in Tracer, and selecting all 4 state_freqs, do these densities make sense given what you know about the model used? answer
- Which parameter is hardest to estimate precisely (i.e. has the broadest HPD credible interval)? answer
- For the parameter you identified in the previous question, would it help to simulate 20000 sites rather than 10000? answer
- What would help in reducing the HPD interval for birth_rate? answer
You may have noticed that our effective sample sizes for the exchangeabilities and state_freqs parameters are pretty low. You no doubt also notice that these are being estimated quite precisely and accurately. What gives? The fact that there is so much information in the data about these parameters is the problem here. The densities for these parameters are very "sharp" (low variance), and proposals that move the values for these parameters away from the optimum by very much fail. Looking at the operator summary table generated after the MCMC analysis finished, you will notice that RevBayes maxed out its tuning parameter for both exchangeabilities and state_freqs at 100. Making this tuning parameter larger results in smaller proposed changes, so if we could set the tuning parameter alpha to, say, 1000 or even 10000, we could achieve better acceptance rates and higher ESSs.
Relaxed clocks
It is safe to assume that a strict molecular clock almost never applies to real data. So, it would be good to allow some flexibility in rates. One common approach is to assume that the rates for each edge are drawn from a lognormal distribution. This is often called an UnCorrelated Lognormal (UCLN) relaxed clock model because the rate for each edge independent of the rate for all other edges (that's the uncorrelated part) and all rates are lognormally distributed. This is to distinguish this approach from correlated relaxed clock models, which assume that rates are to some extent inherited from ancestors, and thus there is autocorrelation across the tree.
Copy your strict.Rev script to a file named relaxed.Rev:
cp strict.Rev relaxed.Rev
Edit relaxed.Rev, replacing the section entitled "Strict clock" with this relaxed clock version:
# Uncorrelated Lognormal relaxed clock
# Add hyperparameters mu and sigma
ucln_mu ~ dnNormal(0.0, 100)
ucln_sigma ~ dnExponential(.01)
moves[nmoves++] = mvSlide(ucln_mu, delta=0.5, tune=true, tuneTarget=0.4, weight=1.0)
moves[nmoves++] = mvSlide(ucln_sigma, delta=0.5, tune=true, tuneTarget=0.4, weight=1.0)
# Create a vector of stochastic nodes representing branch rate parameters
n_branches <- 2*n_taxa - 2
for(i in 1:n_branches) {
branch_rates[i] ~ dnLognormal(ucln_mu, ucln_sigma)
moves[nmoves++] = mvSlide(branch_rates[i], delta=0.5, tune=true, weight=1.0)
}
You'll also need to substitute branch_rates for clock_rate in your dnPhyloCTMC call:
phySeq ~ dnPhyloCTMC(tree=timetree, Q=Q, branchRates=branch_rates, nSites=n_sites, type="DNA")
The model has suddenly gotten a lot more complicated, hasn't it? We now have a rate parameter for every edge in the tree, so we've added 2*20-2=38 more parameters to the model. Each of these edge rate parameters has a lognormal prior, and the 2 parameters of that distribution represent hyperparameters in what is now a hierarchical model, so we've increased the model from 10 parameters (1 clock_rate, 1 birth_rate, 3 state_freqs, 5 exchangeabilities) to 50 parameters (the original 10 plus 38 edge rates and 2 hyperparameters ucln_mean and ucln_stddev).
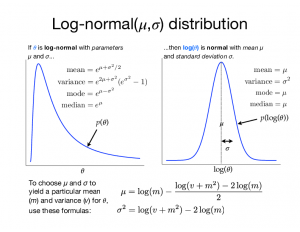
To the right is a figure (click the thumbnail to enlarge) that shows the relationship between the lognormal distribution (left) and the normal distribution (right). The lognormal distribution is strange in that its two parameters (mu and sigma) are not the mean and standard deviation of the lognormally-distributed variable, as you might be led to assume; they are the mean and standard deviation of the log of the lognormally-distributed variable!
You should also edit the monitors section so that the output file names reflect the fact that we're using a relaxed clock now:
# Monitors monitors[nmonitors++] = mnModel(filename = "output/relaxed.log", printgen = 10, separator = TAB) monitors[nmonitors++] = mnFile(filename = "output/relaxed.trees", printgen = 10, timetree) monitors[nmonitors++] = mnScreen(printgen=100)
And don't forget to change the name of the dot file:
mymodel.graph("relaxed.dot", TRUE, "white")
Now run the new model:
rb relaxed.Rev
Review results of the relaxed clock analysis
If you create a plot of your relaxed.dot file using one of the online Graphviz viewers, the increase in model complexity will be made very apparent!
Open the relaxed.log file in Tracer and think about the following questions before peeking at the answer: