Difference between revisions of "Phylogenetics: MrBayes Lab"
Paul Lewis (Talk | contribs) (→Using Tracer to summarize MCMC results) |
|||
| (181 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
{| border="0" | {| border="0" | ||
|- | |- | ||
|rowspan="2" valign="top"|[[Image:Adiantum.png|150px]] | |rowspan="2" valign="top"|[[Image:Adiantum.png|150px]] | ||
| − | |<span style="font-size: x-large">[http://hydrodictyon.eeb.uconn.edu/eebedia/index.php/Phylogenetics:_Syllabus EEB | + | |<span style="font-size: x-large">[http://hydrodictyon.eeb.uconn.edu/eebedia/index.php/Phylogenetics:_Syllabus EEB 5349: Phylogenetics]</span> |
|- | |- | ||
| − | |The goal of this lab exercise is to introduce you to the most important | + | |The goal of this lab exercise is to introduce you to one of the most important computer programs for conducting Bayesian phylogenetic analyses, [http://mrbayes.csit.fsu.edu/ MrBayes]. We will be using MrBayes v3.2.4 x64 on the cluster in this lab. You will also learn how to use the program [http://tree.bio.ed.ac.uk/software/tracer/ Tracer] to analyze MrBayes' output. |
|} | |} | ||
| − | + | == Getting started == | |
| + | == Login to Xanadu == | ||
| − | + | Login to Xanadu and request a machine as usual: | |
| − | + | srun --pty -p mcbstudent --qos=mcbstudent bash | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | Once you are transferred to a free node, type | ||
| + | module load MrBayes/3.2.7 | ||
| − | + | === Create a directory === | |
| − | + | Use the unix mkdir command to create a directory to play in today: | |
| + | mkdir mblab | ||
| − | and | + | === Download and save the data file === |
| − | + | Save the contents of the file [http://hydrodictyon.eeb.uconn.edu/people/plewis/courses/phylogenetics/data/algaemb.nex algaemb.nex] to a file in the mblab folder. One easy way to do this is to cd into the mblab folder, then use the curl command ("Copy URL") to download the file: | |
| + | cd mblab | ||
| + | curl -O http://hydrodictyon.eeb.uconn.edu/people/plewis/courses/phylogenetics/data/algaemb.nex | ||
| − | + | Use nano to look at this file. This is the same 16S data you have used before, but the original file had to be changed to accommodate MrBayes' eccentricities. While MrBayes does a fair job of reading Nexus files, it chokes on certain constructs. The information about what I had to change in order to get it to work is in a comment at the top of the file (this might be helpful in converting other nexus files to work with MrBayes in the future). | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | == Creating a MRBAYES block == | |
| + | The next step is to set up an MCMC analysis. There are three commands in particular that you will need to know about in order to set up a typical run: <tt>lset</tt>, <tt>prset</tt> and <tt>mcmc</tt>. The command <tt>mcmcp</tt> is identical to <tt>mcmc</tt> except that it does not actually start a run. For each of these commands you can obtain online information by typing <tt>help</tt> followed by the command name: for example, <tt>help prset</tt>. Start MrBayes interactively by simply typing <tt>mb</tt> to see how to get help: | ||
| + | mb | ||
| + | This will start MrBayes. At the "MrBayes>" prompt, type <tt>help</tt>: | ||
| + | MrBayes> help | ||
| + | To quit MrBayes, type <tt>quit</tt> at the "MrBayes>" prompt. You will need to quit MrBayes in order to build up the MRBAYES block in your data file. | ||
| − | < | + | Create a MRBAYES block in your Nexus data file. MrBayes does not have a built-in editor, so you will need to use the nano editor to edit the <tt>algaemb.nex</tt> data file. Use Ctrl-/, Ctrl-v to jump to the bottom of the file in nano, then add the following at the ''very bottom'' of the file to begin creating the MRBAYES block: |
| + | begin mrbayes; | ||
| + | set autoclose=yes; | ||
| + | end; | ||
| + | Note that I refer to this block as a MRBAYES block (upper case), but the MrBayes program does not care about case, so using mrbayes (lower case) works just fine. <!-- The <tt>seed=1</tt> and <tt>swapseed=1</tt> statements in the <tt>set</tt> command tell MrBayes to set the pseudorandom number seeds to a particular value. That way you will get the same run I did when I wrote the answers to many of the questions that follow. --> The <tt>autoclose=yes</tt> statement in the <tt>set</tt> command tells MrBayes that we will not want to continue the run beyond the 10,000 generations specified. If you leave this out, MrBayes will ask you whether you wish to continue running the chains after the specified number of generations is finished. | ||
| − | < | + | Add subsequent commands (described below) after the <tt>set</tt> command and before the <tt>end;</tt> line. Note that commands in MrBayes are (intentionally) similar to those in PAUP*, but the differences can be frustrating. For instance, <tt>lset ?</tt> in PAUP* gives you information about the current likelihood settings, but this does not work in MrBayes. Instead, you type <tt>help lset</tt>. Also, the <tt>lset</tt> command in MrBayes has many options not present in PAUP*, and ''vice versa''. |
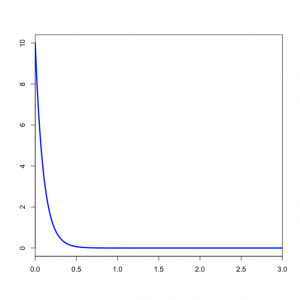
| − | < | + | === Specifying the prior on branch lengths === |
| + | [[Image:Exp10.png|right|thumb|Exponential(10) density function]] | ||
| + | begin mrbayes; | ||
| + | set autoclose=yes; | ||
| + | '''prset brlenspr=unconstrained:exp(10.0);''' | ||
| + | end; | ||
| + | The <tt>prset</tt> command above specifies that branch lengths are to be unconstrained (i.e. a molecular clock is not assumed) and the prior distribution to be applied to each branch length is an exponential distribution with mean 1/10. Note that the value you specify for <tt>unconstrained:exp</tt> is the ''inverse'' of the mean.<br clear="right"/> | ||
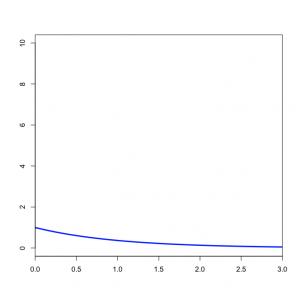
| − | + | === Specifying the prior on the gamma shape parameter === | |
| + | [[Image:Exp1.png|right|thumb|Exponential(1) density function]] | ||
| + | begin mrbayes; | ||
| + | set autoclose=yes; | ||
| + | prset brlenspr=unconstrained:exp(10.0); | ||
| + | '''prset shapepr=exp(1.0);''' | ||
| + | end; | ||
| + | The second <tt>prset</tt> command specifies an exponential distribution with mean 1.0 for the shape parameter of the gamma distribution we will use to model rate heterogeneity. Note that we have not yet told MrBayes that we wish to assume that substitution rates are variable - we will do that using the <tt>lset</tt> command below.<br clear="right"/> | ||
| − | + | === Specifying the prior on kappa === | |
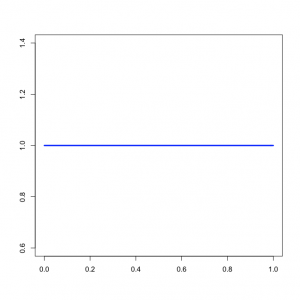
| + | [[Image:Uniform01.png|left|thumb|Beta(1,1) density for transition and transversion rates]] | ||
| − | + | begin mrbayes; | |
| + | set autoclose=yes; | ||
| + | prset brlenspr=unconstrained:exp(10.0); | ||
| + | prset shapepr=exp(1.0); | ||
| + | '''prset tratiopr=beta(1.0,1.0);''' | ||
| + | end; | ||
| + | The command above says to use a Beta(1,1) distribution as the prior for the transition/transversion rate ratio. | ||
| + | <div style="background-color: #ccccff"> | ||
| + | ''Based on what you know about Beta distributions, does it make sense to use a Beta distribution as a prior for the transition/transversion rate ratio?'' {{title|it is strange because tratio ranges from 0 to infinity and the Beta distribution has support only from 0 to 1|answer}} | ||
| + | </div> | ||
| + | Allow me to explain the use of a Beta distribution for tratio as best I can. Recall that the kappa parameter is the ratio <math>\alpha/\beta</math>, where <math>\alpha</math> is the rate of transitions and <math>\beta</math> is the rate of transversions. Rather than allowing you to place a prior directly on the ratio <math>\alpha/\beta</math>, which ranges from 0 to infinity, MrBayes asks you to instead place a joint (Beta) prior on <math>\alpha/(\alpha + \beta)</math> and <math>\beta/(\alpha + \beta)</math>. Here, <math>\alpha/(\alpha + \beta)</math> and <math>\beta/(\alpha + \beta)</math> act like <math>p</math> and <math>1-p</math> in the familiar coin flipping experiment. The reasoning behind this is esoteric, but is the same as the reasoning behind the (now commonplace) use of Dirichlet priors for the GTR relative rates, which is explained nicely in Zwickl, D., and Holder, M. T. 2004. Model parameterization, prior distributions, and the general time-reversible model in Bayesian phylogenetics. Systematic Biology 53(6):877–888. | ||
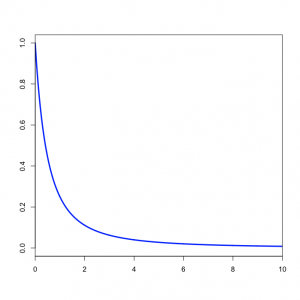
| − | + | [[Image:Exp1ratio.png|right|thumb|BetaPrime(1,1) density for kappa]] | |
| + | You might wonder what the Beta(1,1) distribution (figure on the left) implies about kappa. Transforming the Beta density into the density of <math>\alpha/\beta</math> results in the plot on the right. This density for kappa is very close, but not identical, to an exponential(1) distribution. This is known as the [http://en.wikipedia.org/wiki/Beta_prime_distribution Beta Prime distribution], and has support [0, infinity), which is appropriate for a ratio such as kappa. The Beta Prime distribution is somewhat peculiar, however, when both parameters are 1 (as they are in this case): in this case, the mean is not defined, which is to say that we cannot predict the mean of a sample of kappa values drawn from this distribution. It is not essential for a prior distribution to have a well-defined mean, so even though this is a little weird it nevertheless works pretty well.<br clear="both"/> | ||
| − | + | === Specifying a prior on base frequencies === | |
| + | begin mrbayes; | ||
| + | set autoclose=yes; | ||
| + | prset brlenspr=unconstrained:exp(10.0); | ||
| + | prset shapepr=exp(1.0); | ||
| + | prset tratiopr=beta(1.0,1.0); | ||
| + | '''prset statefreqpr=dirichlet(1.0,1.0,1.0,1.0);''' | ||
| + | end; | ||
| + | The above command states that a flat Dirichlet distribution is to be used for base frequencies. The Dirichlet distribution is like the Beta distribution, except that it is applicable to ''combinations'' of parameters. Like the Beta distribution, the distribution is symmetrical if all the parameters of the distribution are equal, and the distribution is flat if all the parameters of the distribution are equal to 1.0. Using the command above specifies a flat Dirichlet prior, which says that any combination of base frequencies will be given equal prior weight. This means that (0.01, 0.99, 0.0, 0.0) has the same probability density, ''a priori'', as (0.25, 0.25, 0.25, 0.25). If you wanted base frequencies to not stray much from (0.25, 0.25, 0.25, 0.25), you could specify, say, <tt>statefreqpr=dirichlet(10.0,10.0,10.0,10.0)</tt> instead. | ||
| − | + | === The lset command === | |
| + | begin mrbayes; | ||
| + | set autoclose=yes; | ||
| + | prset brlenspr=unconstrained:exp(10.0); | ||
| + | prset shapepr=exp(1.0); | ||
| + | prset tratiopr=beta(1.0,1.0); | ||
| + | prset statefreqpr=dirichlet(1.0,1.0,1.0,1.0); | ||
| + | '''lset nst=2 rates=gamma ngammacat=4;''' | ||
| + | end; | ||
| + | We are finished setting priors now, so the <tt>lset</tt> command above finishes our specification of the model by telling MrBayes that we would like a 2-parameter substitution matrix (i.e. the rate matrix has only two substitution rates, the transition rate and the transversion rate). It also specifies that we would like rates to vary across sites according to a gamma distribution with 4 categories. | ||
| − | + | === Specifying MCMC options === | |
| + | begin mrbayes; | ||
| + | set autoclose=yes; | ||
| + | prset brlenspr=unconstrained:exp(10.0); | ||
| + | prset shapepr=exp(1.0); | ||
| + | prset tratiopr=beta(1.0,1.0); | ||
| + | prset statefreqpr=dirichlet(1.0,1.0,1.0,1.0); | ||
| + | lset nst=2 rates=gamma ngammacat=4; | ||
| + | '''mcmcp ngen=10000 samplefreq=10 printfreq=100 nruns=1 nchains=3 savebrlens=yes;''' | ||
| + | end; | ||
| + | The <tt>mcmcp</tt> command above specifies most of the remaining details of the analysis. | ||
| − | < | + | <tt>ngen=10000</tt> tells MrBayes that its robots should each take 10,000 steps. You should ordinarily use much larger values for <tt>ngen</tt> than this (the default is 1 million steps). We're keeping it small here because we do not have a lot of time and the purpose of this lab is to learn how to use MrBayes, not produce a publishable result. |
| − | < | + | <tt>samplefreq=10</tt> says to only save parameter values and the tree topology every 10 steps. |
| − | < | + | <tt>printfreq=100</tt> says that we would like a progress report every 100 steps. |
| − | < | + | <tt>nruns=1</tt> says to just do one independent run. MrBayes performs two separate analyses by default. |
| − | < | + | <tt>nchains=3</tt> says that we would like to have 2 heated chains running in addition to the cold chain. |
| − | < | + | Finally, <tt>savebrlens=yes</tt> tells MrBayes that we would like it to save branch lengths when it saves the sampled tree topologies. |
| − | < | + | === Specifying an outgroup === |
| − | + | begin mrbayes; | |
| − | + | set autoclose=yes; | |
| − | + | prset brlenspr=unconstrained:exp(10.0); | |
| − | + | prset shapepr=exp(1.0); | |
| − | + | prset tratiopr=beta(1.0,1.0); | |
| − | + | prset statefreqpr=dirichlet(1.0,1.0,1.0,1.0); | |
| − | + | lset nst=2 rates=gamma ngammacat=4; | |
| − | + | mcmcp ngen=10000 samplefreq=10 printfreq=100 nruns=1 nchains=3 savebrlens=yes; | |
| − | + | '''outgroup Anacystis_nidulans;''' | |
| − | + | end; | |
| − | + | The <tt>outgroup</tt> command merely affects the display of trees. It says we want trees to be rooted between the taxon <tt>Anacystis_nidulans</tt> and everything else. | |
| − | + | ||
| − | of | + | == Running MrBayes and interpreting the results == |
| − | and | + | Now save the file and start MrBayes (from within the <tt>mblab</tt> directory) by typing |
| − | + | mb | |
| − | + | Once it starts, type the following at the <tt>MrBayes></tt> prompt | |
| − | + | exe algaemb.nex | |
| − | + | Alternatively, you could both start MrBayes and execute the data file like this: | |
| − | + | mb -i algaemb.nex | |
| − | + | The <tt>-i</tt> tells MrBayes to wait for further commands (i.e. be interactive). | |
| − | + | ||
| − | + | Once MrBayes has finished executing the data file, type the following at the <tt>MrBayes ></tt> prompt: | |
| − | + | mcmc | |
| − | + | This command starts the run. While MrBayes runs, it shows one-line progress reports. The first column is the iteration (generation) number. The next three columns show the log-likelihoods of the separate chains that are running, with the cold chain indicated by square brackets rather than parentheses. The last complete column is a prediction of the time remaining until the run completes. The columns consisting of only -- are simply separators, they have no meaning. | |
| − | + | <div style="background-color: #ccccff"> | |
| − | + | * ''Do you see evidence that the 3 chains are swapping with each other?'' {{title|yes, the square brackets move around indicating swaps have taken place|answer}} | |
| − | + | </div> | |
| − | </ | + | |
| − | + | The section entitled ''Chain swap information:'' reports the number of times each of the three chains attempted to swap with one of the other chains (three values in lower left, below the main diagonal) and the proportion of time such attempts were successful (three values in upper right, above the main diagonal). | |
| − | + | <div style="background-color: #ccccff"> | |
| − | + | * ''How many times did MrBayes attempt to swap chains per generation? Use the information in the lower diagonal of the chain swap information table for this, in conjunction with the number of total generations you specified in the MRBAYES block'' {{title|MrBayes attempts to swap a random pair of chains once per generation, as indicated by the fact that the total number of swap attempts equals the number of generations|answer}} | |
| − | + | </div> | |
| − | + | ||
| − | + | When the run has finished, MrBayes will report (in the section entitled ''Acceptance rates for the moves in the "cold" chain:'') various statistics about the run, such as the percentage of time it was able to accept proposed changes of various sorts. These percentages should, ideally, all be between about 20% and 50%, but as long as they are not extreme (e.g. 1% or 99%) then things went well. Even if there are low acceptance rates for some proposal types, this may not be important if there are other proposal types that operate on the same parameters. For example, note that ExtSPR, ExtTBR, NNI and PrsSPR all operate on Tau, which is the tree topology. As long as these proposals are collectively effective, the fact that one of them is accepting at a very low rate is not of concern. | |
| − | + | <div style="background-color: #ccccff"> | |
| − | + | * ''What explanation could you offer if the acceptance rate was very low, e.g. 1%?'' {{title|proposals are too bold and tend to propose places too far down the hill that are then rejected|answer}} | |
| − | + | * ''What explanation could you offer if the acceptance rate was very high, e.g. 99%?'' {{title|proposals are not bold enough and tend to propose places very close to the current position, which are accepted with high probability because they cannot be very far downhill|answer}} | |
| − | + | </div> | |
| − | + | Below is the acceptance information for my run: | |
| − | + | Acceptance rates for the moves in the "cold" chain: | |
| − | + | With prob. (last 100) chain accepted proposals by move | |
| − | + | 38.3 % ( 32 %) Dirichlet(Tratio) | |
| − | + | 21.7 % ( 18 %) Dirichlet(Pi) | |
| − | + | NA NA Slider(Pi) | |
| − | + | 49.3 % ( 52 %) Multiplier(Alpha) | |
| − | + | 9.0 % ( 14 %) ExtSPR(Tau,V) | |
| − | + | 2.3 % ( 5 %) ExtTBR(Tau,V) | |
| − | + | 10.3 % ( 17 %) NNI(Tau,V) | |
| − | + | 9.8 % ( 8 %) ParsSPR(Tau,V) | |
| − | + | 46.3 % ( 39 %) Multiplier(V) | |
| − | + | 29.8 % ( 28 %) Nodeslider(V) | |
| − | + | 19.6 % ( 22 %) TLMultiplier(V) | |
| − | + | In the above table, 49.3% of proposals to change the gamma shape parameter (denoted Alpha by MrBayes) were accepted. This makes it sounds as if the gamma shape parameter was changed quite often, but to get the full picture, you need to scroll up to the beginning of the output and examine this section: | |
| − | + | The MCMC sampler will use the following moves: | |
| − | + | With prob. Chain will use move | |
| − | + | 2.00 % Dirichlet(Tratio) | |
| − | + | 1.00 % Dirichlet(Pi) | |
| − | + | 1.00 % Slider(Pi) | |
| − | + | 2.00 % Multiplier(Alpha) | |
| − | + | 10.00 % ExtSPR(Tau,V) | |
| − | + | 10.00 % ExtTBR(Tau,V) | |
| − | + | 10.00 % NNI(Tau,V) | |
| − | + | 10.00 % ParsSPR(Tau,V) | |
| − | + | 40.00 % Multiplier(V) | |
| − | + | 10.00 % Nodeslider(V) | |
| − | + | 4.00 % TLMultiplier(V) | |
| − | + | This says that an attempt to change the gamma shape parameter will only be made in 2% of the iterations. | |
| − | + | <div style="background-color: #ccccff"> | |
| − | + | * ''How many times did MrBayes attempt to modify the gamma shape parameter?'' {{title|2% 0f 10000 is 200 times|answer}} | |
| − | + | * ''How many times did MrBayes actually modify the gamma shape parameter?'' {{title|49.3% of 2% 0f 10000 is 99 times|answer}} | |
| − | + | </div> | |
| − | + | ||
| − | + | The fact that MrBayes modified the gamma shape parameter fewer than 100 times out of a run involving 10000 iterations brings up a couple of important points. First, in each iteration, MrBayes chooses a move (i.e. proposal) at random to try. Each move is associated with a "Rel. prob." (relative probability). Using the <tt>showmoves</tt> command shows the following list of moves that were used in this particular analysis: | |
| − | + | 1 -- Move = Dirichlet(Tratio) | |
| − | + | Type = Dirichlet proposal | |
| − | + | Parameter = Tratio [param. 1] (Transition and transversion rates) | |
| − | + | Tuningparam = alpha (Dirichlet parameter) | |
| − | + | alpha = 49.010 [chain 1] | |
| − | + | 49.502 [chain 2] | |
| − | + | 49.502 [chain 3] | |
| − | + | Targetrate = 0.250 | |
| − | + | Rel. prob. = 1.0 | |
| − | + | ||
| − | < | + | 2 -- Move = Dirichlet(Pi) |
| − | + | Type = Dirichlet proposal | |
| − | + | Parameter = Pi [param. 2] (Stationary state frequencies) | |
| − | + | Tuningparam = alpha (Dirichlet parameter) | |
| − | + | alpha = 101.005 [chain 1] | |
| − | + | 101.005 [chain 2] | |
| − | + | 100.000 [chain 3] | |
| − | + | Targetrate = 0.250 | |
| − | + | Rel. prob. = 0.5 | |
| − | + | ||
| − | + | 3 -- Move = Slider(Pi) | |
| − | + | Type = Sliding window | |
| + | Parameter = Pi [param. 2] (Stationary state frequencies) | ||
| + | Tuningparam = delta (Sliding window size) | ||
| + | delta = 0.202 [chain 1] | ||
| + | 0.202 [chain 2] | ||
| + | 0.200 [chain 3] | ||
| + | Targetrate = 0.250 | ||
| + | Rel. prob. = 0.5 | ||
| + | |||
| + | 4 -- Move = Multiplier(Alpha) | ||
| + | Type = Multiplier | ||
| + | Parameter = Alpha [param. 3] (Shape of scaled gamma distribution of site rates) | ||
| + | Tuningparam = lambda (Multiplier tuning parameter) | ||
| + | lambda = 0.827 [chain 1] | ||
| + | 0.827 [chain 2] | ||
| + | 0.819 [chain 3] | ||
| + | Targetrate = 0.250 | ||
| + | Rel. prob. = 1.0 | ||
| + | |||
| + | 5 -- Move = ExtSPR(Tau,V) | ||
| + | Type = Extending SPR | ||
| + | Parameters = Tau [param. 5] (Topology) | ||
| + | V [param. 6] (Branch lengths) | ||
| + | Tuningparam = p_ext (Extension probability) | ||
| + | lambda (Multiplier tuning parameter) | ||
| + | p_ext = 0.500 | ||
| + | lambda = 0.098 | ||
| + | Rel. prob. = 5.0 | ||
| + | |||
| + | 6 -- Move = ExtTBR(Tau,V) | ||
| + | Type = Extending TBR | ||
| + | Parameters = Tau [param. 5] (Topology) | ||
| + | V [param. 6] (Branch lengths) | ||
| + | Tuningparam = p_ext (Extension probability) | ||
| + | lambda (Multiplier tuning parameter) | ||
| + | p_ext = 0.500 | ||
| + | lambda = 0.098 | ||
| + | Rel. prob. = 5.0 | ||
| + | |||
| + | 7 -- Move = NNI(Tau,V) | ||
| + | Type = NNI move | ||
| + | Parameters = Tau [param. 5] (Topology) | ||
| + | V [param. 6] (Branch lengths) | ||
| + | Rel. prob. = 5.0 | ||
| + | |||
| + | 8 -- Move = ParsSPR(Tau,V) | ||
| + | Type = Parsimony-biased SPR | ||
| + | Parameters = Tau [param. 5] (Topology) | ||
| + | V [param. 6] (Branch lengths) | ||
| + | Tuningparam = warp (parsimony warp factor) | ||
| + | lambda (multiplier tuning parameter) | ||
| + | r (reweighting probability) | ||
| + | warp = 0.100 | ||
| + | lambda = 0.098 | ||
| + | r = 0.050 | ||
| + | Rel. prob. = 5.0 | ||
| + | |||
| + | 9 -- Move = Multiplier(V) | ||
| + | Type = Random brlen hit with multiplier | ||
| + | Parameter = V [param. 6] (Branch lengths) | ||
| + | Tuningparam = lambda (Multiplier tuning parameter) | ||
| + | lambda = 2.048 | ||
| + | Targetrate = 0.250 | ||
| + | Rel. prob. = 20.0 | ||
| + | |||
| + | 10 -- Move = Nodeslider(V) | ||
| + | Type = Node slider (uniform on possible positions) | ||
| + | Parameter = V [param. 6] (Branch lengths) | ||
| + | Tuningparam = lambda (Multiplier tuning parameter) | ||
| + | lambda = 0.191 | ||
| + | Rel. prob. = 5.0 | ||
| + | |||
| + | 11 -- Move = TLMultiplier(V) | ||
| + | Type = Whole treelength hit with multiplier | ||
| + | Parameter = V [param. 6] (Branch lengths) | ||
| + | Tuningparam = lambda (Multiplier tuning parameter) | ||
| + | lambda = 1.332 [chain 1] | ||
| + | 1.345 [chain 2] | ||
| + | 1.332 [chain 3] | ||
| + | Targetrate = 0.250 | ||
| + | Rel. prob. = 2.0 | ||
| + | |||
| + | Use 'Showmoves allavailable=yes' to see a list of all available moves | ||
| + | Summing the 11 relative probabilities yields 1 + 0.5 + 0.5 + 1 + 5 + 5 + 5 + 5 + 20 + 5 + 2 = 50. To get the probability of using one of these moves in any particular iteration, MrBayes divides the relative probability for the move by this sum. Thus, move 4, whose job is to update the gamma shape parameter (called Alpha by MrBayes) will be chosen with probability 1/50 = 0.02. This is where the "2.00 % Multiplier(Alpha)" line comes from in the move probability table spit out just before the run started. | ||
| + | |||
| + | Second, note that MrBayes places a lot of emphasis on modifying the tree topology and branch lengths (in this case 94% of proposals), but puts little effort (in this case only 6%) into updating other model parameters. You can change the percent effort for a particular move using the <tt>propset</tt> command. For example, to increase the effort devoted to updating the gamma shape parameter, you could ('''but don't do this now!''') issue the following command either at the MrBayes prompt or in a MRBAYES block: | ||
| + | propset Multiplier(Alpha)$prob=10 | ||
| + | This will change the relative probability of the "Multiplier(Alpha)" move from its default value 1 to the value you specified (10). You can also change tuning parameters for moves using the <tt>propset</tt> command. Before doing that, however, we need to see if the boldness of any moves needs to be changed. | ||
| + | |||
| + | === The sump command === | ||
| + | MrBayes saves information in several files. Only two of these will concern us today. One of them will be called <tt>algaemb.nex.p</tt>. This is the file in which the sampled parameter values were saved. This file is saved as a tab-delimited text file so it is possible to read it into a variety of programs that can be used for summarization or plotting. We will examine this file graphically in a moment, but first let's get MrBayes to summarize its contents for us. | ||
| + | |||
| + | At the MrBayes prompt, type the command <tt>sump</tt>. This will generate a crude graph showing the log-likelihood as a function of time. Note that the log-likelihood starts out low on the left (you started from a random tree, remember), then quickly climbs to a range of values just below -3176. | ||
| + | |||
| + | Below the graph, MrBayes provides the arithmetic mean and harmonic mean of the marginal likelihood. The '''harmonic mean''' has been often used in estimating '''Bayes factors''', which are in turn useful for deciding which among different models fits the data best on average. We will talk about how to use this value in lecture, where you will also get some dire warnings about Bayes factors calculated in this way. | ||
| + | |||
| + | The table at the end is quite useful. It shows the posterior mean, median, variance and 95% credible interval for each parameter in your model based on the samples taken during the run. The credible interval shows the range of values of a parameter that account for the middle 95% of its marginal posterior distribution. If the credible interval for kappa is 3.8 to 6.8, then you can say that there is a 95% chance that kappa is between 3.8 and 6.8 given your data and the assumed model. The parameter TL represents the sum of all the branch lengths. Rather than report every branch length individually, MrBayes just keeps track of their sum. | ||
| + | |||
| + | Look at the output of the sump command and answer these questions: | ||
| + | <div style="background-color: #ccccff"> | ||
| + | * ''What is the total number of samples saved from the posterior distribution?'' {{title|1001|answer}} | ||
| + | * ''How many iterations (generations) did you specify in your MRBAYES block?'' {{title|10000|answer}} | ||
| + | * ''Explain why the two numbers above are different.'' {{title|sampled only every 10th iteration, which yields 1000 samples; the 1 additional sample represents the starting state|answer}} | ||
| + | * ''What proportion of the sampled values did MrBayes automatically exclude as burn-in?'' {{title|25%, as indicated by the statement: Based on a total of 751 samples out of a total of 1001 samples|answer}} | ||
| + | * ''Which value in the parameter column had the largest effective sample size (ESS)?'' {{title|TL|answer}} | ||
| + | * ''Would you conclude from the ESS column that a longer run is necessary?'' {{title|yes, I found that only 2 parameters have ESS values greater than 100|answer}} | ||
| + | </div> | ||
| + | |||
| + | === The sumt command === | ||
| + | Now type the command <tt>sumt</tt>. This will summarize the trees that have been saved in the file <tt>algaemb.nex.t</tt>. | ||
| + | |||
| + | The output of this command includes a bipartition (=split) table, showing posterior probabilities for every split found in any tree sampled during the run. After the bipartition table is shown a majority-rule consensus tree (labeled ''Clade credibility values'') containing all splits that had posterior probability 0.5 or above. | ||
| + | |||
| + | If you chose to save branch lengths (and we did), MrBayes shows a second tree (labeled ''Phylogram'') in which each branch is displayed in such a way that branch lengths are proportional to their posterior mean. MrBayes keeps a running sum of the branch lengths for particular splits it finds in trees as it reads the file <tt>algaemb.nex.t</tt>. Before displaying this tree, it divides the sum for each split by the total number of times it encountered the split to get a simple average branch length for each split. It then draws the tree so that branch lengths are proportional to these mean branch lengths. | ||
| + | |||
| + | Finally, the last thing the <tt>sumt</tt> command does is tell you how many tree topologies are in credible sets of various sizes. For example, in my run, it said that the 99% credible set contained 16 trees. What does this tell us? MrBayes orders tree topologies from most frequent to least frequent (where frequency refers to the number of times they appear in <tt>algaemb.nex.t</tt>). To construct the 99% credible set of trees, it begins by adding the most frequent tree to the set. If that tree accounts for 99% or more of the posterior probability (i.e. at least 99% of all the trees in the <tt>algaemb.nex.t</tt> file have this topology), then MrBayes would say that the 99% credible set contains 1 tree. If the most frequent tree topology was not that frequent, then MrBayes would add the next most frequent tree topology to the set. If the combined posterior probability of both trees was at least 0.99, it would say that the 99% credible set contains 2 trees. In our case, it had to add the top 16 trees to get the total posterior probability up to 99%. | ||
| + | |||
| + | Type <tt>quit</tt> (or just <tt>q</tt>), to quit MrBayes now. | ||
| + | |||
| + | == Using Tracer to summarize MCMC results == | ||
| + | The Java program [http://tree.bio.ed.ac.uk/software/tracer/ Tracer] is very useful for summarizing the results of Bayesian phylogenetic analyses. Tracer was written to accompany the program [http://tree.bio.ed.ac.uk/software/beast/ Beast], but it works well with the output file produced by MrBayes as well. This lab was written using Tracer version 1.6. | ||
| + | |||
| + | To use Tracer on your own computer to view files created on the cluster, you need to get the file on the cluster downloaded to your laptop. Download the file <tt>algaemb.nex.p</tt> (using Cyberduck, FileZilla, Fugu, scp, or whatever has been working). | ||
| + | |||
| + | After starting Tracer, choose ''File > Import Trace File...'' to choose a parameter sample file to display (you can also do this by clicking the + button under the list of trace files in the upper left corner of the main window). Select the <tt>algaemb.nex.p</tt> in your working folder, then click the ''Open'' button to read it. | ||
| + | |||
| + | You should now see 9 rows of values in the table labeled ''Traces'' on the left side of the main window. The first row (''LnL'') is selected by default, and Tracer shows a histogram of log-likelihood values on the right, with summary statistics above the histogram. | ||
| + | |||
| + | A histogram is perhaps not the most useful plot to make with the LnL values. Click the Trace tab to see a trace plot (plot of the log-likelihood through time). | ||
| + | |||
| + | Tracer determines the burn-in period using an undocumented algorithm. You may wish to be more conservative than Tracer. Opinions vary about burn-in. Some Bayesians feel it is important to exclude the first few samples because it is obvious that the chains have not reached stationarity at this point. Other Bayesians feel that if you are worried about the effect of the earliest samples, then you definitely have not run your chains long enough! You might be interested in reading [http://www.stat.umn.edu/~charlie/mcmc/burn.html Charlie Geyer's rant] on burn-in some time. | ||
| + | |||
| + | Because our MrBayes run was just to learn how to run MrBayes and not to do a serious analysis, the trace plot of the log-likelihood will reflect the fact that in this case the burn-in period should be at least 20% of the run! A longer run is also indicated by all the ESS values shown in <span style="color:red; font-weight:bold">red</span> in the Traces panel. Tracer shows an ESS in red if it is less than 200, which it treats as the minimal effective sample size. | ||
| + | <div style="background-color: #ccccff"> | ||
| + | * ''What is the effective sample size for TL? {{title|around 140|answer}} | ||
| + | * ''What did MrBayes report as the effective sample size for TL? {{title|155.18|answer}} | ||
| + | * ''Why is there a difference? Hint: compare the burn-in for both. {{title|Tracer excluded 10% of samples as burn-in, while MrBayes excluded 25%|answer}} | ||
| + | * ''Explain why the ESS reported by MrBayes is higher than that reported by Tracer even though fewer samples were included by MrBayes. {{title|MrBayes cut out all of the initial climb out of randomness, leaving only samples that were much less autocorrelated|answer}} | ||
| + | </div> | ||
| + | |||
| + | '''Before going further!!!''' Change the burn-in used by Tracer from 1000 to 2500 so that the burn-in includes all of the initial climb out of randomness evident in the trace plot of LnL. | ||
| + | |||
| + | Click the Estimates tab again at the top, then click the row labeled kappa on the left. | ||
| + | <div style="background-color: #ccccff"> | ||
| + | * ''What is the posterior mean of kappa?'' {{title|4.8487|answer}} | ||
| + | * ''What is the 95% credible interval for kappa?'' {{title|3.8506 to 5.8384|answer}} | ||
| + | </div> | ||
| + | |||
| + | Click the row labeled ''alpha'' on the left. This is the shape parameter of the gamma distribution governing rates across sites. | ||
| + | |||
| + | <div style="background-color: #ccccff"> | ||
| + | * ''What is the posterior mean of alpha?'' {{title|0.2498|answer}} | ||
| + | * ''What is the 95% credible interval for alpha?'' {{title|0.1663 to 0.3145|answer}} | ||
| + | * ''Is there rate heterogeneity among sites, or are all sites evolving at nearly the same rate?'' {{title|a gamma shape parameter considerably less than 1 indicates substantial rate heterogeneity|answer}} | ||
| + | </div> | ||
| + | |||
| + | Click on the row labeled ''TL'' on the left (the Tree Length). | ||
| + | |||
| + | <div style="background-color: #ccccff"> | ||
| + | * ''What is the posterior mean tree length?'' {{title|0.646|answer}} | ||
| + | * ''What is the mean edge length? (Hint: divide the tree length by the number of edges, which is 2n-3 if n is the number of taxa.) {{title|0.646 divided by 13 equals 0.0496|answer}} | ||
| + | </div> | ||
| + | |||
| + | === Scatterplots of pairs of parameters === | ||
| + | Note that Tracer lets you easily create scatterplots of combinations of parameters. Simply select two parameters (you will have to hold down the Ctrl or Cmd key to select multiple items) and then click on the Joint-Marginal tab. | ||
| + | |||
| + | === Marginal densities === | ||
| + | Try selecting all four base frequencies and then clicking the Marginal Prob Distribution tab. This will show (estimated) marginal probability density plots for all four frequencies at once. Note that KDE is selected underneath the plot in the Display drop-down list. KDE stands for "Kernel Density Estimation" and represents a common non-parametric method for smoothing histograms into estimates of probabilty density functions. | ||
| + | |||
| + | == Running MrBayes with no data == | ||
| + | Why would you want to run MrBayes with no data? Here's a possible reason. You discover by reading the text that results from typing <tt>help prset</tt> that MrBayes assumes, by default, the following branch length prior: <tt>exp(10)</tt>. What does the 10 mean here? Is this an exponential distribution with mean 10 or is 10 the "rate" parameter (a common way to parameterize the exponential distribution)? If 10 is correctly interpreted as the rate parameter, then the mean of the distribution is 1/rate, or 0.1. Even good documentation such as that provided for MrBayes does not explicitly spell out everything you might want to know, but running MrBayes without data can provide answers, at least to questions concerning prior distributions. | ||
| + | |||
| + | Also, it is not possible to place prior distributions directly on some quantities of interest. For example, while you can specify a flat prior on topologies, it is not possible to place a prior on a particular split you are interested in. This is because the prior distribution of splits is '''induced''' by the prior you place on topologies. Running a Bayesian MCMC program without data is a good way to make sure you know what priors you are actually placing on the quantities of interest. | ||
| + | |||
| + | If there is no information in the data, the posterior distribution equals the prior distribution. An MCMC analysis in such cases provides an approximation of the prior. MrBayes makes it easy to run the MCMC analysis without data. (For programs that don't make it easy, simply create a data set containing just one site for which each taxon has missing data.) | ||
| + | |||
| + | Start by deleting the output from the earlier run of the <tt>algaemb.nex</tt> data file: | ||
| + | rm -f algaemb.nex.* | ||
| + | The above command will leave the data file <tt>algaemb.nex</tt> behind, but delete files with names based on the data file but which append other characters to the filename, such as <tt>algaemb.nex.p</tt> and <tt>algaemb.nex.t</tt>. The -f means "force" (i.e. don't ask, just delete). It goes without saying that you should not use <tt>rm -f</tt> if you are tired! | ||
| + | |||
| + | If the only file remaining in the <tt>mblab</tt> directory is the <tt>algaemb.nex</tt> data file, type the following to start the data-free analysis: | ||
| + | mb -i algaemb.nex | ||
| + | MrBayes> mcmc data=no ngen=1000000 samplefreq=100 | ||
| + | Note that I have increased the number of generations to 1 million because the run will go very fast. Sampling every 100th generation will give us a sample of size 10000 to work with. | ||
| + | |||
| + | <div style="background-color: #ccccff"> | ||
| + | * ''Consulting Bayes' formula, what value of the likelihood would cause the posterior to equal the prior?'' {{title|1.0|answer}} | ||
| + | * ''Is this the value that MrBayes reports for the log-likelihood in this case?'' {{title|yes, the log-likelihood is 0.0, which corresponds to a likelihood equal to 1.0|answer}} | ||
| + | </div> | ||
| + | |||
| + | === Checking the shape parameter prior === | ||
| + | Import the output file <tt>algaemb.nex.p</tt> in Tracer. Look first at the histogram of alpha, the shape parameter of the gamma distribution. | ||
| + | |||
| + | <div style="background-color: #ccccff"> | ||
| + | * ''What is the mean you expected for alpha based on the <tt>prset shapepr=exp(1.0)</tt> command in the <tt>blank.nex</tt> file?'' {{title|1.0|answer}} | ||
| + | * ''What is the posterior mean actually estimated by MrBayes (and presented by Tracer)?'' {{title|0.9856|answer}} | ||
| + | * ''An exponential distribution always starts high and approaches zero as you move to the right along the x-axis. The highest point of the exponential density function is 1/mean. If you look at the approximated density plot (click on the ''Marginal Density'' tab), does it appear to approach 1/mean at the value alpha=0.0?'' {{title|yes, but changing Display from KDE to Histogram may make it clearer|answer}} | ||
| + | </div> | ||
| + | |||
| + | === Checking the branch length prior === | ||
| + | Now look at the histogram of TL, the tree length. | ||
| + | |||
| + | <div style="background-color: #ccccff"> | ||
| + | * ''What is the posterior mean of TL, as reported by Tracer?'' {{title|1.2944|answer}} | ||
| + | * ''What value did you expect based on the <tt>prset brlenspr=unconstrained:exp(10)</tt> command?'' {{title|prior mean edge length 0.1 multiplied by 13 edges equals 1.3|answer}} | ||
| + | * ''Does the approximated posterior distribution of TL appear to be an exponential distribution?'' {{title|no, it has a mode to the right of 1 whereas an exponential distribution peaks at 0|answer}} | ||
| + | </div> | ||
| − | + | The second and third questions are a bit tricky, so I'll just give you the explanation. Please make sure this explanation makes sense to you, however, and ask us to explain further if it doesn't make sense. We told MrBayes to place an exponential prior with mean 0.1 on each branch. There are 13 branches in a 8-taxon, unrooted tree. Thus, 13 times 0.1 equals 1.3, which should be close to the posterior mean you obtained for TL. That part is fairly straightforward. | |
| − | + | ||
| − | + | The marginal distribution of TL does not look at all like an exponential distribution, despite the fact that TL should be the sum of 13 exponential distributions. It turns out that the sum of <math>n</math> independent Exponential(<math>\lambda</math>) distributions is a Gamma(<math>n</math>, <math>1/\lambda</math>) distribution. In our case the tree length distribution is a sum of 13 independent Exponential(10) distributions, which equals a Gamma(13, 0.1) distribution. Such a Gamma distribution would have a mean of 1.3 and a peak (mode) at 1.2. If you want to visualize this, fire up R and type the following commands: | |
| − | + | curve(dgamma(theta, shape=13, scale=0.1), from=0, to=2, xname="theta") | |
| − | + | <div style="background-color: #ccccff"> | |
| − | + | * ''How does the Gamma(13, 0.1) density compare to the distribution of TL as shown by Tracer? (Be sure to click the "Marginal Density" tab in Tracer)'' {{title|looks the same|answer}} | |
| − | + | </div> | |
| − | + | ||
| − | + | === Other output files produced by MrBayes === | |
| − | + | That's it for the lab today. You can look at plots of the other parameters if you like. You should also spend some time opening the other output files MrBayes produces in a text editor to make sure you understand what information is saved in these files. Note that some of MrBayes' output files are actually Nexus tree files, which you can open in [http://tree.bio.ed.ac.uk/software/figtree/ FigTree]. For example, <tt>algaemb.nex.t</tt> contains the sampled trees; however, if there are many trees in <tt>algaemb.nex.t</tt>, be prepared for a long wait while FigTree loads the file. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | |||
[[Category:Phylogenetics]] | [[Category:Phylogenetics]] | ||
Latest revision as of 01:34, 9 March 2020

|
EEB 5349: Phylogenetics |
| The goal of this lab exercise is to introduce you to one of the most important computer programs for conducting Bayesian phylogenetic analyses, MrBayes. We will be using MrBayes v3.2.4 x64 on the cluster in this lab. You will also learn how to use the program Tracer to analyze MrBayes' output. |
Contents
Getting started
Login to Xanadu
Login to Xanadu and request a machine as usual:
srun --pty -p mcbstudent --qos=mcbstudent bash
Once you are transferred to a free node, type
module load MrBayes/3.2.7
Create a directory
Use the unix mkdir command to create a directory to play in today:
mkdir mblab
Download and save the data file
Save the contents of the file algaemb.nex to a file in the mblab folder. One easy way to do this is to cd into the mblab folder, then use the curl command ("Copy URL") to download the file:
cd mblab curl -O http://hydrodictyon.eeb.uconn.edu/people/plewis/courses/phylogenetics/data/algaemb.nex
Use nano to look at this file. This is the same 16S data you have used before, but the original file had to be changed to accommodate MrBayes' eccentricities. While MrBayes does a fair job of reading Nexus files, it chokes on certain constructs. The information about what I had to change in order to get it to work is in a comment at the top of the file (this might be helpful in converting other nexus files to work with MrBayes in the future).
Creating a MRBAYES block
The next step is to set up an MCMC analysis. There are three commands in particular that you will need to know about in order to set up a typical run: lset, prset and mcmc. The command mcmcp is identical to mcmc except that it does not actually start a run. For each of these commands you can obtain online information by typing help followed by the command name: for example, help prset. Start MrBayes interactively by simply typing mb to see how to get help:
mb
This will start MrBayes. At the "MrBayes>" prompt, type help:
MrBayes> help
To quit MrBayes, type quit at the "MrBayes>" prompt. You will need to quit MrBayes in order to build up the MRBAYES block in your data file.
Create a MRBAYES block in your Nexus data file. MrBayes does not have a built-in editor, so you will need to use the nano editor to edit the algaemb.nex data file. Use Ctrl-/, Ctrl-v to jump to the bottom of the file in nano, then add the following at the very bottom of the file to begin creating the MRBAYES block:
begin mrbayes; set autoclose=yes; end;
Note that I refer to this block as a MRBAYES block (upper case), but the MrBayes program does not care about case, so using mrbayes (lower case) works just fine. The autoclose=yes statement in the set command tells MrBayes that we will not want to continue the run beyond the 10,000 generations specified. If you leave this out, MrBayes will ask you whether you wish to continue running the chains after the specified number of generations is finished.
Add subsequent commands (described below) after the set command and before the end; line. Note that commands in MrBayes are (intentionally) similar to those in PAUP*, but the differences can be frustrating. For instance, lset ? in PAUP* gives you information about the current likelihood settings, but this does not work in MrBayes. Instead, you type help lset. Also, the lset command in MrBayes has many options not present in PAUP*, and vice versa.
Specifying the prior on branch lengths
begin mrbayes; set autoclose=yes; prset brlenspr=unconstrained:exp(10.0); end;
The prset command above specifies that branch lengths are to be unconstrained (i.e. a molecular clock is not assumed) and the prior distribution to be applied to each branch length is an exponential distribution with mean 1/10. Note that the value you specify for unconstrained:exp is the inverse of the mean.
Specifying the prior on the gamma shape parameter
begin mrbayes; set autoclose=yes; prset brlenspr=unconstrained:exp(10.0); prset shapepr=exp(1.0); end;
The second prset command specifies an exponential distribution with mean 1.0 for the shape parameter of the gamma distribution we will use to model rate heterogeneity. Note that we have not yet told MrBayes that we wish to assume that substitution rates are variable - we will do that using the lset command below.
Specifying the prior on kappa
begin mrbayes; set autoclose=yes; prset brlenspr=unconstrained:exp(10.0); prset shapepr=exp(1.0); prset tratiopr=beta(1.0,1.0); end;
The command above says to use a Beta(1,1) distribution as the prior for the transition/transversion rate ratio.
Based on what you know about Beta distributions, does it make sense to use a Beta distribution as a prior for the transition/transversion rate ratio? answer
Allow me to explain the use of a Beta distribution for tratio as best I can. Recall that the kappa parameter is the ratio 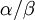 , where
, where 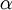 is the rate of transitions and
is the rate of transitions and  is the rate of transversions. Rather than allowing you to place a prior directly on the ratio
is the rate of transversions. Rather than allowing you to place a prior directly on the ratio 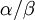 , which ranges from 0 to infinity, MrBayes asks you to instead place a joint (Beta) prior on
, which ranges from 0 to infinity, MrBayes asks you to instead place a joint (Beta) prior on 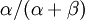 and
and 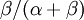 . Here,
. Here, 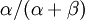 and
and 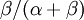 act like
act like 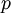 and
and 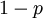 in the familiar coin flipping experiment. The reasoning behind this is esoteric, but is the same as the reasoning behind the (now commonplace) use of Dirichlet priors for the GTR relative rates, which is explained nicely in Zwickl, D., and Holder, M. T. 2004. Model parameterization, prior distributions, and the general time-reversible model in Bayesian phylogenetics. Systematic Biology 53(6):877–888.
in the familiar coin flipping experiment. The reasoning behind this is esoteric, but is the same as the reasoning behind the (now commonplace) use of Dirichlet priors for the GTR relative rates, which is explained nicely in Zwickl, D., and Holder, M. T. 2004. Model parameterization, prior distributions, and the general time-reversible model in Bayesian phylogenetics. Systematic Biology 53(6):877–888.
You might wonder what the Beta(1,1) distribution (figure on the left) implies about kappa. Transforming the Beta density into the density of 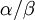 results in the plot on the right. This density for kappa is very close, but not identical, to an exponential(1) distribution. This is known as the Beta Prime distribution, and has support [0, infinity), which is appropriate for a ratio such as kappa. The Beta Prime distribution is somewhat peculiar, however, when both parameters are 1 (as they are in this case): in this case, the mean is not defined, which is to say that we cannot predict the mean of a sample of kappa values drawn from this distribution. It is not essential for a prior distribution to have a well-defined mean, so even though this is a little weird it nevertheless works pretty well.
results in the plot on the right. This density for kappa is very close, but not identical, to an exponential(1) distribution. This is known as the Beta Prime distribution, and has support [0, infinity), which is appropriate for a ratio such as kappa. The Beta Prime distribution is somewhat peculiar, however, when both parameters are 1 (as they are in this case): in this case, the mean is not defined, which is to say that we cannot predict the mean of a sample of kappa values drawn from this distribution. It is not essential for a prior distribution to have a well-defined mean, so even though this is a little weird it nevertheless works pretty well.
Specifying a prior on base frequencies
begin mrbayes; set autoclose=yes; prset brlenspr=unconstrained:exp(10.0); prset shapepr=exp(1.0); prset tratiopr=beta(1.0,1.0); prset statefreqpr=dirichlet(1.0,1.0,1.0,1.0); end;
The above command states that a flat Dirichlet distribution is to be used for base frequencies. The Dirichlet distribution is like the Beta distribution, except that it is applicable to combinations of parameters. Like the Beta distribution, the distribution is symmetrical if all the parameters of the distribution are equal, and the distribution is flat if all the parameters of the distribution are equal to 1.0. Using the command above specifies a flat Dirichlet prior, which says that any combination of base frequencies will be given equal prior weight. This means that (0.01, 0.99, 0.0, 0.0) has the same probability density, a priori, as (0.25, 0.25, 0.25, 0.25). If you wanted base frequencies to not stray much from (0.25, 0.25, 0.25, 0.25), you could specify, say, statefreqpr=dirichlet(10.0,10.0,10.0,10.0) instead.
The lset command
begin mrbayes; set autoclose=yes; prset brlenspr=unconstrained:exp(10.0); prset shapepr=exp(1.0); prset tratiopr=beta(1.0,1.0); prset statefreqpr=dirichlet(1.0,1.0,1.0,1.0); lset nst=2 rates=gamma ngammacat=4; end;
We are finished setting priors now, so the lset command above finishes our specification of the model by telling MrBayes that we would like a 2-parameter substitution matrix (i.e. the rate matrix has only two substitution rates, the transition rate and the transversion rate). It also specifies that we would like rates to vary across sites according to a gamma distribution with 4 categories.
Specifying MCMC options
begin mrbayes; set autoclose=yes; prset brlenspr=unconstrained:exp(10.0); prset shapepr=exp(1.0); prset tratiopr=beta(1.0,1.0); prset statefreqpr=dirichlet(1.0,1.0,1.0,1.0); lset nst=2 rates=gamma ngammacat=4; mcmcp ngen=10000 samplefreq=10 printfreq=100 nruns=1 nchains=3 savebrlens=yes; end;
The mcmcp command above specifies most of the remaining details of the analysis.
ngen=10000 tells MrBayes that its robots should each take 10,000 steps. You should ordinarily use much larger values for ngen than this (the default is 1 million steps). We're keeping it small here because we do not have a lot of time and the purpose of this lab is to learn how to use MrBayes, not produce a publishable result.
samplefreq=10 says to only save parameter values and the tree topology every 10 steps.
printfreq=100 says that we would like a progress report every 100 steps.
nruns=1 says to just do one independent run. MrBayes performs two separate analyses by default.
nchains=3 says that we would like to have 2 heated chains running in addition to the cold chain.
Finally, savebrlens=yes tells MrBayes that we would like it to save branch lengths when it saves the sampled tree topologies.
Specifying an outgroup
begin mrbayes; set autoclose=yes; prset brlenspr=unconstrained:exp(10.0); prset shapepr=exp(1.0); prset tratiopr=beta(1.0,1.0); prset statefreqpr=dirichlet(1.0,1.0,1.0,1.0); lset nst=2 rates=gamma ngammacat=4; mcmcp ngen=10000 samplefreq=10 printfreq=100 nruns=1 nchains=3 savebrlens=yes; outgroup Anacystis_nidulans; end;
The outgroup command merely affects the display of trees. It says we want trees to be rooted between the taxon Anacystis_nidulans and everything else.
Running MrBayes and interpreting the results
Now save the file and start MrBayes (from within the mblab directory) by typing
mb
Once it starts, type the following at the MrBayes> prompt
exe algaemb.nex
Alternatively, you could both start MrBayes and execute the data file like this:
mb -i algaemb.nex
The -i tells MrBayes to wait for further commands (i.e. be interactive).
Once MrBayes has finished executing the data file, type the following at the MrBayes > prompt:
mcmc
This command starts the run. While MrBayes runs, it shows one-line progress reports. The first column is the iteration (generation) number. The next three columns show the log-likelihoods of the separate chains that are running, with the cold chain indicated by square brackets rather than parentheses. The last complete column is a prediction of the time remaining until the run completes. The columns consisting of only -- are simply separators, they have no meaning.
- Do you see evidence that the 3 chains are swapping with each other? answer
The section entitled Chain swap information: reports the number of times each of the three chains attempted to swap with one of the other chains (three values in lower left, below the main diagonal) and the proportion of time such attempts were successful (three values in upper right, above the main diagonal).
- How many times did MrBayes attempt to swap chains per generation? Use the information in the lower diagonal of the chain swap information table for this, in conjunction with the number of total generations you specified in the MRBAYES block answer
When the run has finished, MrBayes will report (in the section entitled Acceptance rates for the moves in the "cold" chain:) various statistics about the run, such as the percentage of time it was able to accept proposed changes of various sorts. These percentages should, ideally, all be between about 20% and 50%, but as long as they are not extreme (e.g. 1% or 99%) then things went well. Even if there are low acceptance rates for some proposal types, this may not be important if there are other proposal types that operate on the same parameters. For example, note that ExtSPR, ExtTBR, NNI and PrsSPR all operate on Tau, which is the tree topology. As long as these proposals are collectively effective, the fact that one of them is accepting at a very low rate is not of concern.
- What explanation could you offer if the acceptance rate was very low, e.g. 1%? answer
- What explanation could you offer if the acceptance rate was very high, e.g. 99%? answer
Below is the acceptance information for my run:
Acceptance rates for the moves in the "cold" chain:
With prob. (last 100) chain accepted proposals by move
38.3 % ( 32 %) Dirichlet(Tratio)
21.7 % ( 18 %) Dirichlet(Pi)
NA NA Slider(Pi)
49.3 % ( 52 %) Multiplier(Alpha)
9.0 % ( 14 %) ExtSPR(Tau,V)
2.3 % ( 5 %) ExtTBR(Tau,V)
10.3 % ( 17 %) NNI(Tau,V)
9.8 % ( 8 %) ParsSPR(Tau,V)
46.3 % ( 39 %) Multiplier(V)
29.8 % ( 28 %) Nodeslider(V)
19.6 % ( 22 %) TLMultiplier(V)
In the above table, 49.3% of proposals to change the gamma shape parameter (denoted Alpha by MrBayes) were accepted. This makes it sounds as if the gamma shape parameter was changed quite often, but to get the full picture, you need to scroll up to the beginning of the output and examine this section:
The MCMC sampler will use the following moves:
With prob. Chain will use move
2.00 % Dirichlet(Tratio)
1.00 % Dirichlet(Pi)
1.00 % Slider(Pi)
2.00 % Multiplier(Alpha)
10.00 % ExtSPR(Tau,V)
10.00 % ExtTBR(Tau,V)
10.00 % NNI(Tau,V)
10.00 % ParsSPR(Tau,V)
40.00 % Multiplier(V)
10.00 % Nodeslider(V)
4.00 % TLMultiplier(V)
This says that an attempt to change the gamma shape parameter will only be made in 2% of the iterations.
- How many times did MrBayes attempt to modify the gamma shape parameter? answer
- How many times did MrBayes actually modify the gamma shape parameter? answer
The fact that MrBayes modified the gamma shape parameter fewer than 100 times out of a run involving 10000 iterations brings up a couple of important points. First, in each iteration, MrBayes chooses a move (i.e. proposal) at random to try. Each move is associated with a "Rel. prob." (relative probability). Using the showmoves command shows the following list of moves that were used in this particular analysis:
1 -- Move = Dirichlet(Tratio)
Type = Dirichlet proposal
Parameter = Tratio [param. 1] (Transition and transversion rates)
Tuningparam = alpha (Dirichlet parameter)
alpha = 49.010 [chain 1]
49.502 [chain 2]
49.502 [chain 3]
Targetrate = 0.250
Rel. prob. = 1.0
2 -- Move = Dirichlet(Pi)
Type = Dirichlet proposal
Parameter = Pi [param. 2] (Stationary state frequencies)
Tuningparam = alpha (Dirichlet parameter)
alpha = 101.005 [chain 1]
101.005 [chain 2]
100.000 [chain 3]
Targetrate = 0.250
Rel. prob. = 0.5
3 -- Move = Slider(Pi)
Type = Sliding window
Parameter = Pi [param. 2] (Stationary state frequencies)
Tuningparam = delta (Sliding window size)
delta = 0.202 [chain 1]
0.202 [chain 2]
0.200 [chain 3]
Targetrate = 0.250
Rel. prob. = 0.5
4 -- Move = Multiplier(Alpha)
Type = Multiplier
Parameter = Alpha [param. 3] (Shape of scaled gamma distribution of site rates)
Tuningparam = lambda (Multiplier tuning parameter)
lambda = 0.827 [chain 1]
0.827 [chain 2]
0.819 [chain 3]
Targetrate = 0.250
Rel. prob. = 1.0
5 -- Move = ExtSPR(Tau,V)
Type = Extending SPR
Parameters = Tau [param. 5] (Topology)
V [param. 6] (Branch lengths)
Tuningparam = p_ext (Extension probability)
lambda (Multiplier tuning parameter)
p_ext = 0.500
lambda = 0.098
Rel. prob. = 5.0
6 -- Move = ExtTBR(Tau,V)
Type = Extending TBR
Parameters = Tau [param. 5] (Topology)
V [param. 6] (Branch lengths)
Tuningparam = p_ext (Extension probability)
lambda (Multiplier tuning parameter)
p_ext = 0.500
lambda = 0.098
Rel. prob. = 5.0
7 -- Move = NNI(Tau,V)
Type = NNI move
Parameters = Tau [param. 5] (Topology)
V [param. 6] (Branch lengths)
Rel. prob. = 5.0
8 -- Move = ParsSPR(Tau,V)
Type = Parsimony-biased SPR
Parameters = Tau [param. 5] (Topology)
V [param. 6] (Branch lengths)
Tuningparam = warp (parsimony warp factor)
lambda (multiplier tuning parameter)
r (reweighting probability)
warp = 0.100
lambda = 0.098
r = 0.050
Rel. prob. = 5.0
9 -- Move = Multiplier(V)
Type = Random brlen hit with multiplier
Parameter = V [param. 6] (Branch lengths)
Tuningparam = lambda (Multiplier tuning parameter)
lambda = 2.048
Targetrate = 0.250
Rel. prob. = 20.0
10 -- Move = Nodeslider(V)
Type = Node slider (uniform on possible positions)
Parameter = V [param. 6] (Branch lengths)
Tuningparam = lambda (Multiplier tuning parameter)
lambda = 0.191
Rel. prob. = 5.0
11 -- Move = TLMultiplier(V)
Type = Whole treelength hit with multiplier
Parameter = V [param. 6] (Branch lengths)
Tuningparam = lambda (Multiplier tuning parameter)
lambda = 1.332 [chain 1]
1.345 [chain 2]
1.332 [chain 3]
Targetrate = 0.250
Rel. prob. = 2.0
Use 'Showmoves allavailable=yes' to see a list of all available moves
Summing the 11 relative probabilities yields 1 + 0.5 + 0.5 + 1 + 5 + 5 + 5 + 5 + 20 + 5 + 2 = 50. To get the probability of using one of these moves in any particular iteration, MrBayes divides the relative probability for the move by this sum. Thus, move 4, whose job is to update the gamma shape parameter (called Alpha by MrBayes) will be chosen with probability 1/50 = 0.02. This is where the "2.00 % Multiplier(Alpha)" line comes from in the move probability table spit out just before the run started.
Second, note that MrBayes places a lot of emphasis on modifying the tree topology and branch lengths (in this case 94% of proposals), but puts little effort (in this case only 6%) into updating other model parameters. You can change the percent effort for a particular move using the propset command. For example, to increase the effort devoted to updating the gamma shape parameter, you could (but don't do this now!) issue the following command either at the MrBayes prompt or in a MRBAYES block:
propset Multiplier(Alpha)$prob=10
This will change the relative probability of the "Multiplier(Alpha)" move from its default value 1 to the value you specified (10). You can also change tuning parameters for moves using the propset command. Before doing that, however, we need to see if the boldness of any moves needs to be changed.
The sump command
MrBayes saves information in several files. Only two of these will concern us today. One of them will be called algaemb.nex.p. This is the file in which the sampled parameter values were saved. This file is saved as a tab-delimited text file so it is possible to read it into a variety of programs that can be used for summarization or plotting. We will examine this file graphically in a moment, but first let's get MrBayes to summarize its contents for us.
At the MrBayes prompt, type the command sump. This will generate a crude graph showing the log-likelihood as a function of time. Note that the log-likelihood starts out low on the left (you started from a random tree, remember), then quickly climbs to a range of values just below -3176.
Below the graph, MrBayes provides the arithmetic mean and harmonic mean of the marginal likelihood. The harmonic mean has been often used in estimating Bayes factors, which are in turn useful for deciding which among different models fits the data best on average. We will talk about how to use this value in lecture, where you will also get some dire warnings about Bayes factors calculated in this way.
The table at the end is quite useful. It shows the posterior mean, median, variance and 95% credible interval for each parameter in your model based on the samples taken during the run. The credible interval shows the range of values of a parameter that account for the middle 95% of its marginal posterior distribution. If the credible interval for kappa is 3.8 to 6.8, then you can say that there is a 95% chance that kappa is between 3.8 and 6.8 given your data and the assumed model. The parameter TL represents the sum of all the branch lengths. Rather than report every branch length individually, MrBayes just keeps track of their sum.
Look at the output of the sump command and answer these questions:
- What is the total number of samples saved from the posterior distribution? answer
- How many iterations (generations) did you specify in your MRBAYES block? answer
- Explain why the two numbers above are different. answer
- What proportion of the sampled values did MrBayes automatically exclude as burn-in? answer
- Which value in the parameter column had the largest effective sample size (ESS)? answer
- Would you conclude from the ESS column that a longer run is necessary? answer
The sumt command
Now type the command sumt. This will summarize the trees that have been saved in the file algaemb.nex.t.
The output of this command includes a bipartition (=split) table, showing posterior probabilities for every split found in any tree sampled during the run. After the bipartition table is shown a majority-rule consensus tree (labeled Clade credibility values) containing all splits that had posterior probability 0.5 or above.
If you chose to save branch lengths (and we did), MrBayes shows a second tree (labeled Phylogram) in which each branch is displayed in such a way that branch lengths are proportional to their posterior mean. MrBayes keeps a running sum of the branch lengths for particular splits it finds in trees as it reads the file algaemb.nex.t. Before displaying this tree, it divides the sum for each split by the total number of times it encountered the split to get a simple average branch length for each split. It then draws the tree so that branch lengths are proportional to these mean branch lengths.
Finally, the last thing the sumt command does is tell you how many tree topologies are in credible sets of various sizes. For example, in my run, it said that the 99% credible set contained 16 trees. What does this tell us? MrBayes orders tree topologies from most frequent to least frequent (where frequency refers to the number of times they appear in algaemb.nex.t). To construct the 99% credible set of trees, it begins by adding the most frequent tree to the set. If that tree accounts for 99% or more of the posterior probability (i.e. at least 99% of all the trees in the algaemb.nex.t file have this topology), then MrBayes would say that the 99% credible set contains 1 tree. If the most frequent tree topology was not that frequent, then MrBayes would add the next most frequent tree topology to the set. If the combined posterior probability of both trees was at least 0.99, it would say that the 99% credible set contains 2 trees. In our case, it had to add the top 16 trees to get the total posterior probability up to 99%.
Type quit (or just q), to quit MrBayes now.
Using Tracer to summarize MCMC results
The Java program Tracer is very useful for summarizing the results of Bayesian phylogenetic analyses. Tracer was written to accompany the program Beast, but it works well with the output file produced by MrBayes as well. This lab was written using Tracer version 1.6.
To use Tracer on your own computer to view files created on the cluster, you need to get the file on the cluster downloaded to your laptop. Download the file algaemb.nex.p (using Cyberduck, FileZilla, Fugu, scp, or whatever has been working).
After starting Tracer, choose File > Import Trace File... to choose a parameter sample file to display (you can also do this by clicking the + button under the list of trace files in the upper left corner of the main window). Select the algaemb.nex.p in your working folder, then click the Open button to read it.
You should now see 9 rows of values in the table labeled Traces on the left side of the main window. The first row (LnL) is selected by default, and Tracer shows a histogram of log-likelihood values on the right, with summary statistics above the histogram.
A histogram is perhaps not the most useful plot to make with the LnL values. Click the Trace tab to see a trace plot (plot of the log-likelihood through time).
Tracer determines the burn-in period using an undocumented algorithm. You may wish to be more conservative than Tracer. Opinions vary about burn-in. Some Bayesians feel it is important to exclude the first few samples because it is obvious that the chains have not reached stationarity at this point. Other Bayesians feel that if you are worried about the effect of the earliest samples, then you definitely have not run your chains long enough! You might be interested in reading Charlie Geyer's rant on burn-in some time.
Because our MrBayes run was just to learn how to run MrBayes and not to do a serious analysis, the trace plot of the log-likelihood will reflect the fact that in this case the burn-in period should be at least 20% of the run! A longer run is also indicated by all the ESS values shown in red in the Traces panel. Tracer shows an ESS in red if it is less than 200, which it treats as the minimal effective sample size.
- What is the effective sample size for TL? answer
- What did MrBayes report as the effective sample size for TL? answer
- Why is there a difference? Hint: compare the burn-in for both. answer
- Explain why the ESS reported by MrBayes is higher than that reported by Tracer even though fewer samples were included by MrBayes. answer
Before going further!!! Change the burn-in used by Tracer from 1000 to 2500 so that the burn-in includes all of the initial climb out of randomness evident in the trace plot of LnL.
Click the Estimates tab again at the top, then click the row labeled kappa on the left.
- What is the posterior mean of kappa? answer
- What is the 95% credible interval for kappa? answer
Click the row labeled alpha on the left. This is the shape parameter of the gamma distribution governing rates across sites.
- What is the posterior mean of alpha? answer
- What is the 95% credible interval for alpha? answer
- Is there rate heterogeneity among sites, or are all sites evolving at nearly the same rate? answer
Click on the row labeled TL on the left (the Tree Length).
- What is the posterior mean tree length? answer
- What is the mean edge length? (Hint: divide the tree length by the number of edges, which is 2n-3 if n is the number of taxa.) answer
Scatterplots of pairs of parameters
Note that Tracer lets you easily create scatterplots of combinations of parameters. Simply select two parameters (you will have to hold down the Ctrl or Cmd key to select multiple items) and then click on the Joint-Marginal tab.
Marginal densities
Try selecting all four base frequencies and then clicking the Marginal Prob Distribution tab. This will show (estimated) marginal probability density plots for all four frequencies at once. Note that KDE is selected underneath the plot in the Display drop-down list. KDE stands for "Kernel Density Estimation" and represents a common non-parametric method for smoothing histograms into estimates of probabilty density functions.
Running MrBayes with no data
Why would you want to run MrBayes with no data? Here's a possible reason. You discover by reading the text that results from typing help prset that MrBayes assumes, by default, the following branch length prior: exp(10). What does the 10 mean here? Is this an exponential distribution with mean 10 or is 10 the "rate" parameter (a common way to parameterize the exponential distribution)? If 10 is correctly interpreted as the rate parameter, then the mean of the distribution is 1/rate, or 0.1. Even good documentation such as that provided for MrBayes does not explicitly spell out everything you might want to know, but running MrBayes without data can provide answers, at least to questions concerning prior distributions.
Also, it is not possible to place prior distributions directly on some quantities of interest. For example, while you can specify a flat prior on topologies, it is not possible to place a prior on a particular split you are interested in. This is because the prior distribution of splits is induced by the prior you place on topologies. Running a Bayesian MCMC program without data is a good way to make sure you know what priors you are actually placing on the quantities of interest.
If there is no information in the data, the posterior distribution equals the prior distribution. An MCMC analysis in such cases provides an approximation of the prior. MrBayes makes it easy to run the MCMC analysis without data. (For programs that don't make it easy, simply create a data set containing just one site for which each taxon has missing data.)
Start by deleting the output from the earlier run of the algaemb.nex data file:
rm -f algaemb.nex.*
The above command will leave the data file algaemb.nex behind, but delete files with names based on the data file but which append other characters to the filename, such as algaemb.nex.p and algaemb.nex.t. The -f means "force" (i.e. don't ask, just delete). It goes without saying that you should not use rm -f if you are tired!
If the only file remaining in the mblab directory is the algaemb.nex data file, type the following to start the data-free analysis:
mb -i algaemb.nex MrBayes> mcmc data=no ngen=1000000 samplefreq=100
Note that I have increased the number of generations to 1 million because the run will go very fast. Sampling every 100th generation will give us a sample of size 10000 to work with.
- Consulting Bayes' formula, what value of the likelihood would cause the posterior to equal the prior? answer
- Is this the value that MrBayes reports for the log-likelihood in this case? answer
Checking the shape parameter prior
Import the output file algaemb.nex.p in Tracer. Look first at the histogram of alpha, the shape parameter of the gamma distribution.
- What is the mean you expected for alpha based on the prset shapepr=exp(1.0) command in the blank.nex file? answer
- What is the posterior mean actually estimated by MrBayes (and presented by Tracer)? answer
- An exponential distribution always starts high and approaches zero as you move to the right along the x-axis. The highest point of the exponential density function is 1/mean. If you look at the approximated density plot (click on the Marginal Density tab), does it appear to approach 1/mean at the value alpha=0.0? answer
Checking the branch length prior
Now look at the histogram of TL, the tree length.
- What is the posterior mean of TL, as reported by Tracer? answer
- What value did you expect based on the prset brlenspr=unconstrained:exp(10) command? answer
- Does the approximated posterior distribution of TL appear to be an exponential distribution? answer
The second and third questions are a bit tricky, so I'll just give you the explanation. Please make sure this explanation makes sense to you, however, and ask us to explain further if it doesn't make sense. We told MrBayes to place an exponential prior with mean 0.1 on each branch. There are 13 branches in a 8-taxon, unrooted tree. Thus, 13 times 0.1 equals 1.3, which should be close to the posterior mean you obtained for TL. That part is fairly straightforward.
The marginal distribution of TL does not look at all like an exponential distribution, despite the fact that TL should be the sum of 13 exponential distributions. It turns out that the sum of 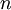 independent Exponential(
independent Exponential(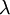 ) distributions is a Gamma(
) distributions is a Gamma(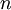 ,
, 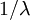 ) distribution. In our case the tree length distribution is a sum of 13 independent Exponential(10) distributions, which equals a Gamma(13, 0.1) distribution. Such a Gamma distribution would have a mean of 1.3 and a peak (mode) at 1.2. If you want to visualize this, fire up R and type the following commands:
) distribution. In our case the tree length distribution is a sum of 13 independent Exponential(10) distributions, which equals a Gamma(13, 0.1) distribution. Such a Gamma distribution would have a mean of 1.3 and a peak (mode) at 1.2. If you want to visualize this, fire up R and type the following commands:
curve(dgamma(theta, shape=13, scale=0.1), from=0, to=2, xname="theta")
- How does the Gamma(13, 0.1) density compare to the distribution of TL as shown by Tracer? (Be sure to click the "Marginal Density" tab in Tracer) answer
Other output files produced by MrBayes
That's it for the lab today. You can look at plots of the other parameters if you like. You should also spend some time opening the other output files MrBayes produces in a text editor to make sure you understand what information is saved in these files. Note that some of MrBayes' output files are actually Nexus tree files, which you can open in FigTree. For example, algaemb.nex.t contains the sampled trees; however, if there are many trees in algaemb.nex.t, be prepared for a long wait while FigTree loads the file.