Difference between revisions of "Phylogenetics: Large Scale Maximum Likelihood Analyses"
(→Comparing GARLI, RaxML, and FastTree) |
(→Comparing GARLI, RaxML, and FastTree) |
||
| Line 251: | Line 251: | ||
#nexus | #nexus | ||
| − | begin trees; | + | begin paup; |
| + | exe rbcL50.nex; | ||
| + | end; | ||
| + | |||
| + | begin trees; | ||
utree garli = (...); | utree garli = (...); | ||
utree raxml = (...); | utree raxml = (...); | ||
| Line 258: | Line 262: | ||
begin paup; | begin paup; | ||
| − | |||
set criterion=likelihood; | set criterion=likelihood; | ||
lset nst=6 rmatrix=estimate rates=gamma shape=estimate; | lset nst=6 rmatrix=estimate rates=gamma shape=estimate; | ||
Revision as of 14:12, 21 February 2011

|
EEB 349: Phylogenetics |
| This lab explores two programs (GARLI and RAxML) designed specifically for maximum likelihood analyses on a large scale (hundreds of taxa). If there is time, we will try a third option, FastTree, which is designed for ML analyses of hundreds of thousands of sequences. |
Contents
- 1 The data
- 2 Part A: Starting a GARLI run on the cluster
- 3 Part B: Starting a RAxML run on the cluster
- 4 Part C: Running FastTree on the cluster
- 5 Comparing GARLI, RaxML, and FastTree
The data
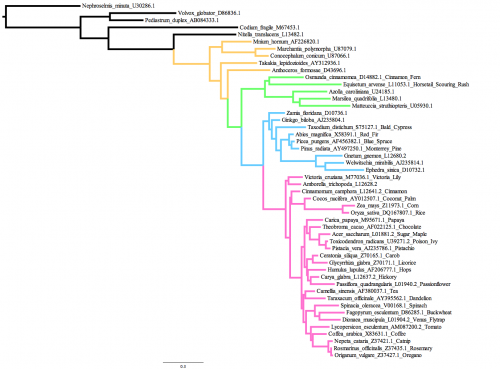
The data that we will use today comprises 50 rbcL sequences from various green plants: 5 green algae, 5 bryophytes (mosses, liverworts, hornworts), 5 ferns, 9 gymnosperms, and 26 angiosperms (flowering plants; including some you will recognize such as oregano, coffee, tea, spinach, poison ivy, rice and chocolate). The tree on the right shows the tree I obtained from GARLI using the GY94 codon model, color-coded so that bryophytes are yellow, ferns are green, gymnosperms are blue and angiosperms are pink. This history of green plants shows several key innovations: embryos are what gave the first bryophytes an edge over aquatic algae on land; branched sporophytes and vascular tissues allowed the first ferns to grow taller and disperse more spores compared to their bryophyte ancestors; seeds and pollen were the big inventions that led to the rise of gymnosperms; and of course flowers allowed efficient pollination by insects and led the the diversification of the angiosperms.Part A: Starting a GARLI run on the cluster
GARLI (Genetic Algorithm for Rapid Likelihood Inference) is a program written by Derrick Zwickl for estimating the phylogeny using maximum likelihood, and is currently one of the best programs to use if you have a large problem (i.e. many taxa). GARLI now (as of version 1.0) gives you considerable choice in substitution models: GTR[+I][+G] or codon models for nucleotides, plus several choices for amino acids. The genetic algorithm (or GA, for short) search strategy used by GARLI is like other heuristic search strategies in that it cannot guarantee that the optimal tree will be found. Thus, as with all heuristic searches, it is a good idea to run GARLI several times (using different pseudorandom number seeds) to see if there is any variation in the estimated tree. By default, GARLI will conduct two independent searches. If you have a multicore processor (newer Intel-based Macs and PCs are duo-core), GARLI can take advantage of this and use all of your CPUs simultaneously.
Today you will run GARLI on the cluster for a dataset with 50 taxa. This is not a particularly large problem, but has the advantage that you will be able to analyze it several times using both GARLI and RAxML within a lab period. Instead of each of us running GARLI several times, we will each run it once and compare notes at the end of the lab.
Preparing the GARLI control file
Like many programs, GARLI uses a control file to specify the settings it will use during a run. Most of the default settings are fine, but you will need to change a few of them before running GARLI.
Obtain a copy of the control file
The first step is to obtain a copy of the GARLI default control file. Go to the GARLI download page and download a version of GARLI appropriate for your platform (Mac or Windows). For now, the only reason you are downloading GARLI is to obtain a copy of the default control file. However, because GARLI is multithreaded, you may find that it is faster to run it on your multi-core laptop than on the cluster. Running on the cluster has advantages, however, even if it is slower. For one, if you have a truly large data set, using the cluster means that your laptop is not tied up for hours.
Once you have downloaded and unpacked GARLI on your computer, copy the garli.conf.nuc.defaultSettings to a file named simply garli.conf and open it in your text editor.
Editing garli.conf
You will only need to change four lines.
Specify the data file name (note the capital L)
datafname = rbcL50.nex
Specify the prefix for output files
ofprefix = 50
The ofprefix is used by GARLI to begin the name of all output files. I usually use something different than the data file name here. That way, if you eventually want to delete all of the various files that GARLI creates, you can just say rm -f 50* without wiping out your data file as well! (Sounds like the voice of experience, doesn't it?!)
Specify no invariable sites
invariantsites = none
This will cause GARLI to use the GTR+G model rather than the GTR+I+G model, which will facilitate comparisons with RAxML.
Do only one search replicate
searchreps = 1
Save the garli.conf file when you have made these changes.
The tip of the GARLI iceberg
As you can see from the number of entries in the control file, we are not going to learn all there is to know about GARLI in one lab session. One major omission is any discussion about bootstrapping, which is very easy to do in GARLI: just set bootstrapreps to some number other than 0 (e.g. 100) in your garli.conf file. I encourage you to download and read the excellent GARLI manual, especially if you want to use amino acid or codon models.
Log into the cluster
Log into the cluster using the command:
ssh bbcxsrv1.biotech.uconn.edu
Go back to the Phylogenetics: Bioinformatics Cluster lab if you've forgotten some details.
Create a folder and a script for the run
Create a directory named garlirun inside your home directory and use your favorite file transfer method (scp, psftp, Fugu, FileZilla, etc.) to get garli.conf into that directory.
Now download the data file into the garlirun directory:
curl http://hydrodictyon.eeb.uconn.edu/eeb5349/rbcL50.nex > rbcL50.nex
Finally, create the script file you will hand to the qsub command to start the run. Use the pico (a.k.a. nano) editor to create a file named gogarli in your home directory with the following contents:
#$ -o junk.txt -j y cd $HOME/garlirun garli garli.conf
Submit the job
Here is the command to start the job:
qsub gogarli
You should issue this command from your home directory, or where ever you saved the gogarli file.
Check progress every few minutes using the qstat command. This run will take about 10 minutes. If you get bored, you can cd into the garlirun directory and use this command to see the tail end of the log file that GARLI creates automatically:
tail 50.log00.log
The tail command is like the cat command except that it only shows you the last few lines of the file (which often is just what you need).
Files produced by GARLI
After your run finishes, you should find these files in your garlirun folder. Download them to your laptop and view them to answer the questions:
50.screen.log
This file saves the output that would have been displayed had you been running GARLI on your laptop.
50.log00.log
This file shows the best log-likelihood at periodic intervals throughout the run. It would be useful if you wanted to plot the progress of the run either as a function of time or generation.
50.best.tre
This is a NEXUS tree file that can be opened in FigTree, TreeView, PAUP*, or a number of other phylogenetic programs. Try using FigTree to open it. The best place to root it is on the branch leading to Nephroselmis. In FigTree, click this branch and use the Reroot tool to change the rooting. I also find that trees look better if you click the Order nodes checkbox, which is inside the Trees tab on the left side panel of FigTree.
Part B: Starting a RAxML run on the cluster
Another excellent ML program for large problems is RAxML, written by Alexandros Stamatakis. This program is exceptionally fast, and has been used to estimate maximum likelihood trees for 25,000 taxa! Let's run RAxML on the same data as GARLI and compare results.
Preparing the data file
While GARLI reads NEXUS files, RAxML uses a simpler format. It is easy to use the pico editor to make the necessary changes, however. First, make a copy of your rbcL50.nex file:
cp rbcL50.nex rbcL50.dat
Open rbcL50.dat in pico and use Ctrl-k repeatedly to remove these initial lines:
#nexus begin data; dimensions ntax=50 nchar=1314; format datatype=dna gap=- missing=?; matrix
Add a new first line to the file that looks like this:
50 1314
Now use the down arrow to go to the end of the file and remove the last two lines:
; end;
Save the file using Ctrl-x and you are ready to run RAxML!
The tip of the RAxML iceberg
As with GARLI, RAxML is full of features that we will not have time to explore today. The manual does a nice job of explaining all the features so I recommend reading it if you use RAxML for your own data.
Running RAxML on the cluster
Hopefully, you have created the rbcL50.dat file in your garlirun directory. If not, go ahead and move it there. Then return to your home directory and use pico to create a gorax script file that contains the following:
#$ -o junk2.txt -j y cd $HOME/garlirun raxml -p 13579 -N 1 -e 0.00001 -m GTRMIX -s rbcL50.dat -n BASIC
You'll note that this is similar to the gogarli script we created earlier, but it is worth discussing each line before submitting the run to the cluster.
The first line is the same except that we specified junk2.txt rather than junk.txt (this is so that our RAxML run will not try to write to the same file as our GARLI run).
The second line is identical to the second line of our gogarli script. You could, of course, sequester the RAxML results in a different directory if you wanted, but it is safe to use the same folder because none of the RAxML output files will have exactly the same name as any of the GARLI output files.
The third line requires the most explanation. First, RAxML does not use a control file like GARLI, so all options must be specified on the command line when it is invoked. Let's take each option in turn:
- -p 13579 provides a pseudorandom number seed to RAxML to use when it generates its starting tree (the p presumably stands for parsimony, which is the optimality criterion it uses to obtain a starting tree). It is a good idea to specify some number here so that you have the option of exactly recreating the analysis later.
- -N 1 tells RAxML to just perform one search replicate.
- -e 0.00001 sets the precision with which model parameters will be estimated. RAxML will search for better combinations of parameter values until it fails to increase the log-likelihood by more than this amount. Ordinarily, the default value (0.1) is sufficient, but we are making RAxML work harder so that the results are more comparable to GARLI, which does a fairly thorough final parameter optimization.
- -m GTRMIX tells RAxML to use the GTR+CAT model for the search, then to switch to the GTR+G for final optimization of parameters (so that the likelihood is comparable to that produced by other programs).
- -s rbcL50.dat provides the name of the data file.
- -n BASIC supplies a suffix to be appended to all output file names
Start the run by entering this from your home directory (or where ever your gorax file is located):
qsub gorax
Bootstrapping with RAxML
After your first RAxML run finishes (probably within 2 minutes), start a second, longer run to perform bootstrapping. Modify your gorax file as follows:
#$ -o junk2.txt -j y cd $HOME/garlirun # raxml -p 13579 -N 1 -e 0.00001 -m GTRMIX -s rbcL50.dat -n BASIC raxml -f a -x 12345 -p 13579 -N 100 -m GTRCAT -s rbcL50.dat -n FULL
Note that I've used a # character to comment out our previous raxml line. (Feel free to simply delete that line if you wish.) Go ahead and start this run using qsub. This one will take longer, but not as long as you might expect (about 10 minutes). It will conduct a bootstrap analysis involving 100 bootstrap replicates (-N 100) using the GTR+CAT model. The -x 12345 specifies a starting seed for the bootstrap resampling. For every 5 bootstrap replicates performed, RAxML will climb uphill on the original dataset starting from the tree estimated for that bootstrap replicate. This provides a series of searches for the maximum likelihood tree starting from different, but reasonable, starting trees. The -f a on the command line sets up this combination of bootstrapping and ML searching.
Files produced by RAxML
RAxML_info.FULL
This file contains some basic information about the run. Use this file to answer these questions:
RAxML_bestTree.FULL
This file holds the best tree found. It is not a NEXUS tree file, but simply a tree description; however, FigTree is able to open such files.
RAxML_bootstrap.FULL
This file holds the trees resulting from bootstrapping (also not NEXUS format; one tree description per line). These trees do not have branch lengths. You can open this file in FigTree and use the arrow buttons to move from one to the next.
RAxML_bipartitions.FULL
This file contains the best tree with bootstrap support values embedded in the tree description. Load this tree into FigTree. FigTree will ask you what name you want to use for the support values. Pick a name such as "bootstraps" and click Ok. Once the tree is visible, check Node labels on the left, chooose "bootstraps" (or whatever you named them) from the Display list, and increase the font size so you can see it (ok, you are probably young enough that you can still see the numbers without magnification!).
Part C: Running FastTree on the cluster
FastTree is probably your best option if you have truly huge data sets. RaxML and/or GARLI are probably better options (more accurate) for data sets of only hundreds to thousands of taxa, but if you have hundreds of thousands of sequences, GARLI and RaxML will be slower than FastTree. FastTree is provided ready to run for Windows and Linux, but for Macs, you will need to compile it yourself. The cluster we've been using is composed of Macs, so your first step will be to download and compile FastTree. There is a lot of phylogenetic software out there, and much of it is available as source code only. Thus, this provides an opportunity to learn how to compile such programs yourself if you do not already know how to do this.
Downloading FastTree
Begin by downloading the FastTree.c file to your home directory on bbcxsrv1.biotech.uconn.edu:
curl http://www.microbesonline.org/fasttree/FastTree.c > FastTree.c
The curl command copies a file from a web site to your terminal, and we are using the operator > to redirect the output of curl to a file named FastTree.c.
About compilers
To create an executable file, you need to run a C compiler. Compilers translate source code (in this case written in the computer language C) to binary (i.e. machine language). The Gnu compiler gcc is ubiquitous on all platforms except Windows, and it is present on the cluster as well. To check, type
which gcc
The which gcc command shows you what command (computer programs are called commands in unix), if any, would be run if you typed gcc at the unix prompt. The fact that the which gcc command yields "/usr/bin/gcc" means that the gcc program exists. Try typing which doofus to see what response is provided when a program does not exist.
If you want to try this on your Mac, you may need to install the Developer Tools in order to get gcc (try the which trick to see if it is already available on your mac). To compile under Windows, you will need to install either the Intel compiler (which will cost you something) or Windows Visual Studio Express (free). For today, we will just compile on the cluster, which is normally what you will want to do because that will allow you to run your analyses on the cluster rather than tying up your own computer.
Compiling FastTree.c
To compile, follow the directions on the FastTree web site by typing the following at the prompt:
gcc -lm -O3 -finline-functions -funroll-loops -Wall -o FastTree FastTree.c
While gcc is working, read the following breakdown of the options we've given it:
- -lm tells gcc that it should link in the math library (this will be necessary for any program that does anything remotely resembling math, such as using the log or exp functions)
- -O3 tells gcc to use the highest level of optimization (more than -O1 or -O2, for example)
- -finline-functions tells gcc to try to inline as many functions as possible. Inlining a function involves replacing calls to the function with the code for that function, which saves the overhead of making the function call (small but adds up if a function is called many times)
- -funroll-loops tells gcc to unrolll for loops if possible. Unrolling a loop means replacing a loop over, say, the four bases with four separate pieces of code, one for each of the four bases. This allows compiler optimizations that would not otherwise be possible.
- -Wall tells gcc to show all warnings
- -o FastTree tells gcc to name the resulting executable file "FastTree"
- FastTree.c tells gcc that the source code is in the file "FastTree.c"
Running FastTree
If you have created FastTree in your home directory, create a new directory named ftree and move the FastTree executable into it. Also, copy the rbcL50.dat file into the fasttree directory as well. cd $HOME mkdir ftree mv FastTree ftree cp garlirun/rbcL50.dat ftree
Now create a qsub script named goftree using pico:
#$ -o junk3.txt -j y cd $HOME/ftree ./FastTree -gtr -nt rbcL50.dat > output.txt
Before you run this file, there are a few points you should note. Note first the "./" before the name of the executable file. This is often necessary when you are invoking a program you compiled yourself. The problem is that the system does not look in the current directory for programs! Try typing which FastTree while inside the ftree directory for example; the system will respond "FastTree: Command not found." -- it doesn't see the FastTree executable even though it is right under its nose! By prefacing the name of the executable by "./", you are explicitly telling it to use the FastTree executable inside the current directory. Try typing this now just to check that this works:
./FastTree
FastTree will run, but since you haven't given it anything to work on, it simply spits out a list of command line options.
Go ahead and submit your qsub script:
qsub goftree
Comparing GARLI, RaxML, and FastTree
To compare the three programs, use the pico editor to create a tree file named combined.nex containing a trees block with the best tree from all three programs and a paup block to compute the likelihoods of these three trees under the GTR+G model. Here I've simply inserted elipses (...) as placeholders for the actual tree descriptions. It may be easier to construct this tree file on your own laptop and then upload it again to the cluster; use whichever approach is most convenient for you.
#nexus begin paup; exe rbcL50.nex; end;
begin trees;
utree garli = (...);
utree raxml = (...);
utree fasttree = (...);
end;
begin paup;
set criterion=likelihood;
lset nst=6 rmatrix=estimate rates=gamma shape=estimate;
lscores all;
end;