Difference between revisions of "Phylogenetics: BEAST Lab"
Paul Lewis (Talk | contribs) (→Adding a calibration) |
Paul Lewis (Talk | contribs) (→Adding a calibration) |
||
| Line 197: | Line 197: | ||
The analyses we performed above all provide relative divergence times under either a strict or relaxed clock, but normally the purpose of a divergence time analysis is to obtain absolute divergence times (in thousands or millions of years) using fossils to calibrate the molecular clock. | The analyses we performed above all provide relative divergence times under either a strict or relaxed clock, but normally the purpose of a divergence time analysis is to obtain absolute divergence times (in thousands or millions of years) using fossils to calibrate the molecular clock. | ||
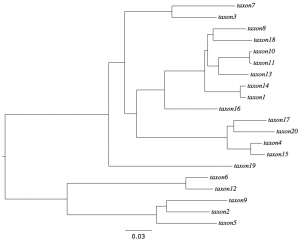
| − | On the right is the tree used to simulate the ''lognormal.nex'' data. | + | [[File:LognormalModelTree.png|thumb|right|Model tree used to simulate data under lognormal relaxed clock]]On the right is the tree used to simulate the ''lognormal.nex'' data. Let's pretend that we have a 100 million year old fossil that we know is in the clade of 5 taxa at the bottom of the tree (comprising taxa 2, 5, 6, 9, and 12). Call this Clade A just to have a label for it. This means that the clade arose started diversifying some time before the age of the fossil. We will assume that the fossil and its date imply that the age of the most recent common ancestor of Clade A occurred, with 95% probability, between 100 and 130 million years ago. |
| + | |||
| + | Start by setting up BEAUTi as you did for the ''lognormal.nex'' data. If you still have BEAUTi open, then it is already set up and ready to go. | ||
| + | |||
| + | Click on the + symbol at the bottom of the ''Priors'' panel. That will allow you to define a calibration prior. Use the buttons provided to include taxa 2, 5, 6, 9, and 12 in a taxon set labeled ''Clade A''. Choose an Exponential prior with ''Mean'' 10 and ''Offset'' 100. Note that the diagram on the right shows that the 95% Quantile is 130; this means that 95% of the distribution is between 100 and 130. We will need to remember that we've specified the distribution in terms of millions of years (i.e. we entered 100 for the offset, not 100,000,000, which is what we would have needed to enter had we wanted times to be in years rather than millions of years. | ||
| + | |||
| + | Not quite done yet... | ||
== Further study == | == Further study == | ||
Revision as of 11:40, 14 April 2016

|
EEB 5349: Phylogenetics |
| In this lab you will learn how to use the program BEAST, and its companion BEAUTi, written by Alexei Drummond, Andrew Rambaut, Marc Suchard, Remco Bouckaert, and numerous other contributors. BEAST specializes in estimating divergence times under uncorrelated relaxed clock models, estimating species trees using a model that accounts for the independent coalescent history of each gene tree, and Bayesian skyline analyses that estimate population growth or decline through time. While BEAST itself is written in C and Java and has no user interface, the companion program BEAUTi has a nice graphical user interface that allows you to create an XML data file that is consulted by BEAST when it runs. |
In this lab, you will use BEAST to estimate divergence times. Rather than analyze a real data set, you will simulated a data set in which you know the true values of all parameters, and then see how to obtain estimates of those parameters in BEAST.
Contents
- 1 Login to the UConn Bioinformatics cluster
- 2 Download BEAUTi and BEAST
- 3 Analysis of strict-clock simulated data
- 3.1 Setting up the Site Model
- 3.2 Setting up the Clock Model
- 3.3 Setting up the Priors
- 3.4 Setting up MCMC
- 3.5 Saving the XML file
- 3.6 Running BEAST on the cluster
- 3.7 Using Tracer to examine the parameter estimates
- 3.8 Using TreeAnnotator and FigTree to examine the tree and divergence time credible intervals
- 4 Analysis of relaxed-clock simulated data
- 5 Further study
Login to the UConn Bioinformatics cluster
Once you login, type
qlogin
to obtain a session on a currently unused node.
Download BEAUTi and BEAST
While we will be using BEAST on the cluster, you will need to download the software locally in order to get BEAUTi, which cannot be easily run remotely on the cluster because of its interactive GUI (Graphical User Interface).
Download the version of BEAST 2.4.0 from the BEAST web site. You will also need FigTree and Tracer (but you should already have those).
BEAST and BEAUti form a pair. BEAUti is a user-friendly, graphical Java application that can be used to create the data file that is then fed to BEAST, which is neither user-friendly nor graphical, but is nevertheless also written in Java. Thus, to do anything with BEAST, you will need a working Java runtime environment. You will actually use the cluster to run BEAST, but you will need to run BEAUti on your own laptop. If you have trouble getting Java running on your laptop, pair with another student until you have an XML file. At that point, you can use your own computer to interact with the cluster.
Analysis of strict-clock simulated data
Download the file simyule.nex to your hard drive and import it into BEAUTi using the File > Import alignment menu command. An entry called simyule should appear in the Partitions section.
The tree from which these data were simulated was itself simulated using the Yule model. Here are some facts about the true tree:
| Number of leaves (species) | 20 |
| Per-lineage speciation rate | 10 |
| Tree length (sum of all edge times) | 1.77537 |
| Tree height (length of path from root to any tip) | 0.279743 |
| log(Yule model probability density) | 25.9954 |
The last line requires some explanation. Simulating a Yule tree requires choosing n-1 times (where n is the number of tips) and, at each step, deciding which of the current lineages will speciate after the chosen sojourn time. The probability density of a Yule tree thus involves the product of n-1 Exponential(lambda*k) densities and n-1 Discrete Uniform(k) probabilities, where k is the number of lineages at each step. The value reported in the table above is the log of this probability density evaluated using the times actually chosen for this particular tree.
Sequence data (10000 sites) was simulated using this Yule tree. Here are some facts about the true substitution model:
| Model | HKY85 |
| Gamma rate heterogeneity shape | 0.5 |
| Equilibrium frequency for A | 0.3 |
| Equilibrium frequency for C | 0.2 |
| Equilibrium frequency for G | 0.2 |
| Equilibrium frequency for T | 0.3 |
| Kappa (trs./trv. rate ratio) | 4.0 |
Setting up the Site Model
We will begin by estimating the parameters of the substitution model that we know was used to generate these data.
Click on the Site Model tab in BEAUTi and set Gamma Category Count to 4, the model to HKY, and check estimate for Shape and Kappa. The Frequencies drop-down list should be set to Estimated, Proportion Invariant should remain fixed to 0.0, and Substitution Rate should be fixed at 1.0.
Setting up the Clock Model
Click on the Clock Model tab in BEAUTi. We will leave everything here at the default values (Strict Clock with Clock.rate fixed to 1). Again, we know the truth, and the truth was that all 10000 sites were generated under a strict clock, with expected number of substitutions equal to the Yule branch lengths (which is why we keep clock rate set at 1, which says that a branch length of 1 means 1 expected substitution per site).
Setting up the Priors
Click on the Priors tab in BEAUTi and set up the priors as follows.
Tree
Keep this set to Yule Model and be sure estimate is checked for Birth Diff Rate (but leave Conditional On Root unchecked)
birthRate
Set this to an Exponential distribution with mean 100 (do not check estimate). An Exponential distribution with mean 100 has variance 100 squared (10000), so this prior should not argue very strongly with the likelihood about the value of the Yule per-lineage speciation rate.
gammaShape
Set this to an Exponential distribution with mean 100 (do not check estimate)
kappa
Set this to an Exponential distribution with mean 100 (do not check estimate)
Setting up MCMC
Click on the MCMC tab in BEAUTi and set the Chain Length to 300000, leave Store Every set to -1, and set Pre Burnin to 10000.
Do NOT check the box at the bottom labeled Sample From Prior
The chain length of 300,000 is way to short, but we can't afford to wait for the default 10,000,000 or we will not be able to finish this in lab. As you will see, 300K MCMC steps is long enough to get most estimates reasonably close to their true values.
Saving the XML file
You are now ready to save the XML file that BEAST will use. To do this, choose File > Save As from the main menu and save the file as simyule.xml in a directory of your choosing. Upload this file to your home directory on the cluster.
Running BEAST on the cluster
I am assuming that you are now logged into the cluster and have issued a qlogin command to acquire a free node. (You should also be able to see your simyule.xml file in your home directory when you type the ls command.)
The version of BEAST that was installed on the cluster was a little out of date, so I installed the latest version (2.4.0 at the time I wrote this) in the directory /common/phylogenetics. To further complicate things, this version of BEAST requires a more recent version of Java than we had available on the cluster, so in that same directory is a freshly downloaded Java Runtime Environment. To make all this work, we have to set an environmental variable so that the default version of Java is not used when we run BEAST.
export PATH="/common/phylogenetics/jre1.8.0_77/bin:$PATH"
This command puts the latest Java Runtime Environment's bin directory at the head of the PATH, where the PATH is a list of directories that the system will search when looking for a program (e.g. java).
You should now be able to start BEAST as follows:
java -jar /common/phylogenetics/beast.jar simyule.xml
Using Tracer to examine the parameter estimates
Once you are finished, bring the log file back to your local computer and open it in Tracer. Use the Estimates tab in Tracer to answer these questions.
- What is the true tree height? answer Does the estimated 95% credible interval for tree height include the true value? answer
- What is the true log Yule model probability density? answer Does the estimated 95% credible interval for Yule model include the true value? answer
- What is the true Yule model per-lineage speciation rate? answer Does the estimated 95% credible interval for birthRate include the true value? answer
- Were the true values of any substitution model parameters outside their respective 95% credible intervals? answer
- If the answer to the last question was "yes", can you explain why the estimate is off by looking at the ESS column and/or the trace plot? answer
Using TreeAnnotator and FigTree to examine the tree and divergence time credible intervals
On your local computer, start the TreeAnnotator program that was downloaded as part of BEAST2. Choose the file simyule.trees for Input Tree File and specify simyulemcc.tre as Output File and press the Run button. After TreeAnnotator finishes, open the file simyulemcc.tre in FigTree and do the following steps after the tree has loaded:
- In the Trees section on the left, check Order nodes (ordering should be increasing)
- Click Node Bars
- Uncheck Scale Bar
- Check Scale Axis and check Reverse axis after opening up the Scale Axis section
Now choose File > New from the main FigTree menu to create a new, blank window. Copy the tree description below and paste it into this window (this is the true tree for comparison):
((((taxon7:0.07448,taxon3:0.07448):0.05612,(((((taxon8:0.04474,taxon18:0.04474):0.00447,((taxon10:0.00318,taxon11:0.00318):0.03218,taxon13:0.03536):0.01384):0.00412,(taxon14:0.00616,taxon1:0.00616):0.04717):0.03522,taxon16:0.08855):0.03100,((taxon17:0.04256,taxon20:0.04256):0.00894,(taxon4:0.01598,taxon15:0.01598):0.03552):0.06805):0.01104):0.02259,taxon19:0.15318):0.12656,((taxon6:0.02920,taxon12:0.02920):0.16953,((taxon9:0.05268,taxon2:0.05268):0.01422,taxon5:0.06690):0.13183):0.08101)
Set up this tree the same way you set up the estimated tree (except checking Node Bars will have no effect this time).
- Does the estimated tree have the correct topology? answer
- Which divergences are associated with the most uncertainty? answer
Analysis of relaxed-clock simulated data
Let's do the exercise again with data that does not conform to a strict molecular clock.
Download the file lognormal.nex to your hard drive and import it into BEAUTi using the File > Import alignment menu command.
The tree and substitution model used to simulate the data in lognormal.nex were identical to the tree and model used to simulate the data in simyule.nex. What's different is that the branch lengths of the tree were modified before the sequences were simulated. There are 38 edges in the tree, so I drew 38 relative rates from a Lognormal(-0.0196104, 0.198042) distribution and multiplied each edge length by one of these rates.
- What was the mean relative rate given that they were independently sampled from a Lognormal distribution with mu = -0.0196104 and sigma = 0.198042? answer (hint: see formula for mean on the Lognormal distribution Wikipedia page)
- What was the true standard deviation of relative rates? answer
If you want to see the effect of multiplying edge lengths by relative rates, copy the tree description below and paste it into a FigTree window:
((((taxon7:0.0719125,taxon3:0.0508014):0.0536504,(((((taxon8:0.0373785,taxon18:0.0443175):0.00624889,((taxon10:0.00242665,taxon11:0.00243614):0.035207,taxon13:0.0342883):0.0125615):0.00328008,(taxon14:0.00630138,taxon1:0.0063153):0.0404656):0.0456252,taxon16:0.0600948):0.0318929,((taxon17:0.0370861,taxon20:0.0462936):0.00773252,(taxon4:0.0126342,taxon15:0.0163993):0.0270225):0.102974):0.0133684):0.0183383,taxon19:0.140667):0.117726,((taxon6:0.0255848,taxon12:0.0313106):0.134908,((taxon9:0.068912,taxon2:0.0489807):0.0116809,taxon5:0.0687154):0.101499):0.0705456)
Setting up the Clock Model
After importing the lognormal.nex data into BEAUTi, set up the Site Model as before and then click on the Clock Model tab. This time choose Relaxed Clock Log Normal but keep Clock.rate fixed to 1.
Setting up the Priors
Set up the priors as before, but you will note that a new parameter has been added: ucldStdev. Set the prior for this parameter to the same Exponential(100) prior distribution we have used for all the other parameters.
Setting up MCMC
Let's do more sampling than before. In the MCMC tab, set Chain Length to 1 million and Pre Burnin to 100,000. Save the file as lognormal.xml, copy it to the cluster, and run it as you did for simyule.xml.
- What was the estimated variance (and credible interval) of relative rates? answer
- What was the true variance of relative rates? answer
Adding a calibration
The analyses we performed above all provide relative divergence times under either a strict or relaxed clock, but normally the purpose of a divergence time analysis is to obtain absolute divergence times (in thousands or millions of years) using fossils to calibrate the molecular clock.
On the right is the tree used to simulate the lognormal.nex data. Let's pretend that we have a 100 million year old fossil that we know is in the clade of 5 taxa at the bottom of the tree (comprising taxa 2, 5, 6, 9, and 12). Call this Clade A just to have a label for it. This means that the clade arose started diversifying some time before the age of the fossil. We will assume that the fossil and its date imply that the age of the most recent common ancestor of Clade A occurred, with 95% probability, between 100 and 130 million years ago.Start by setting up BEAUTi as you did for the lognormal.nex data. If you still have BEAUTi open, then it is already set up and ready to go.
Click on the + symbol at the bottom of the Priors panel. That will allow you to define a calibration prior. Use the buttons provided to include taxa 2, 5, 6, 9, and 12 in a taxon set labeled Clade A. Choose an Exponential prior with Mean 10 and Offset 100. Note that the diagram on the right shows that the 95% Quantile is 130; this means that 95% of the distribution is between 100 and 130. We will need to remember that we've specified the distribution in terms of millions of years (i.e. we entered 100 for the offset, not 100,000,000, which is what we would have needed to enter had we wanted times to be in years rather than millions of years.
Not quite done yet...
Further study
For an tutorial involving analysis of real data, you may wish to explore Tracy Heath's Fossilzed Birth-Death Tutorial.